2XHM
 
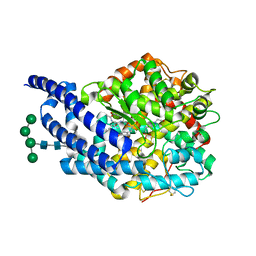 | | Crystal structure of AnCE-K26 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ANGIOTENSIN CONVERTING ENZYME, ... | | Authors: | Akif, M, Ntai, I, Sturrock, E.D, Isaac, R.E, Bachmann, B.O, Acharya, K.R. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of a Phosphonotripeptide K-26 in Complex with Angiotensin Converting Enzyme Homologue (Ance) from Drosophila Melanogaster.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
5D6T
 
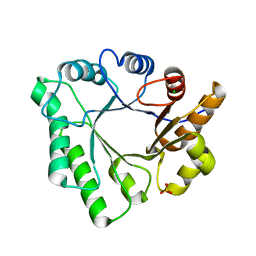 | | Crystal Structure of Aspergillus clavatus Sph3 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, SPHERULIN-4, ... | | Authors: | Bamford, N.C, Little, D.J, Howell, P.L. | | Deposit date: | 2015-08-12 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Sph3 Is a Glycoside Hydrolase Required for the Biosynthesis of Galactosaminogalactan in Aspergillus fumigatus.
J.Biol.Chem., 290, 2015
|
|
8X83
 
 | |
4ZA5
 
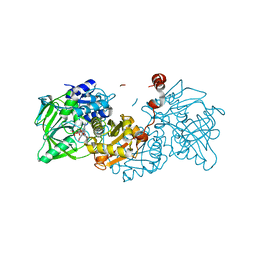 | | Structure of A. niger Fdc1 with the prenylated-flavin cofactor in the iminium and ketimine forms. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 1-deoxy-5-O-phosphono-1-[(10aR)-2,2,3,4-tetramethyl-8,10-dioxo-1,2,8,9,10,10a-hexahydro-6H-indeno[1,7-ef]pyrimido[4,5-b][1,4]diazepin-6-yl]-D-ribitol, Fdc1, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
6F0B
 
 | |
8X84
 
 | |
7CYV
 
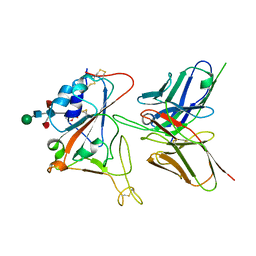 | | Crystal structure of FD20, a neutralizing single-chain variable fragment (scFv) in complex with SARS-CoV-2 Spike receptor-binding domain (RBD) | | Descriptor: | Spike protein S1, The heavy chain variable region of the scFv FD20,The light chain variable region of the scFv FD20, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y, Li, T, Lai, Y, Cai, H, Yao, H, Li, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody.
Embo Mol Med, 13, 2021
|
|
8R2H
 
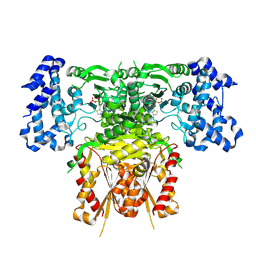 | | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Plasmodium falciparum | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Gawriljuk, V.O, Godoy, A.S, Oerlemans, R, Groves, M.R. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase DXPS from Plasmodium falciparum reveals a distinct N-terminal domain.
Nat Commun, 15, 2024
|
|
5F96
 
 | |
6B3P
 
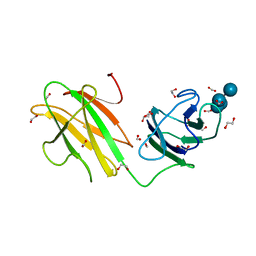 | | Crystal structure of CBMbc (family CBM26) from Eubacterium rectale Amy13K in Complex with Maltoheptaose | | Descriptor: | 1,2-ETHANEDIOL, Amy13K, FORMIC ACID, ... | | Authors: | Cockburn, D.W, Wawrzak, Z, Perez Medina, K, Koropatkin, N.M. | | Deposit date: | 2017-09-22 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Novel carbohydrate binding modules in the surface anchored alpha-amylase of Eubacterium rectale provide a molecular rationale for the range of starches used by this organism in the human gut.
Mol. Microbiol., 107, 2018
|
|
6SWR
 
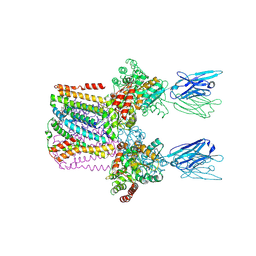 | | Crystal structure of the lysosomal potassium channel MtTMEM175 T38A mutant soaked with zinc | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody, Maltose/maltodextrin-binding periplasmic protein,Maltodextrin-binding protein,Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2019-09-23 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K + channels.
Elife, 9, 2020
|
|
6GLD
 
 | |
5FSG
 
 | |
4RFN
 
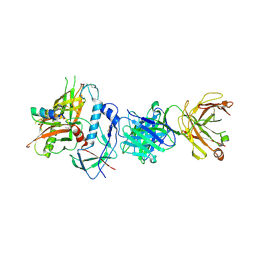 | | Crystal structure of ADCC-potent Rhesus macaque ANTIBODY JR4 in complex with HIV-1 CLADE A/E GP120 and M48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB HEAVY CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, FAB LIGHT CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
8D6K
 
 | | Sco GlgEI-V279S in complex with cyclohexyl carbasugar | | Descriptor: | (1R,4S,5S,6R)-4-(cyclohexylamino)-5,6-dihydroxy-2-(hydroxymethyl)cyclohex-2-en-1-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, DI(HYDROXYETHYL)ETHER | | Authors: | Jayasinghe, T.J, Ronning, D.R. | | Deposit date: | 2022-06-06 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Synthesis of C 7 /C 8 -cyclitols and C 7 N-aminocyclitols from maltose and X-ray crystal structure of Streptomyces coelicolor GlgEI V279S in a complex with an amylostatin GXG-like derivative.
Front Chem, 10, 2022
|
|
2X8Y
 
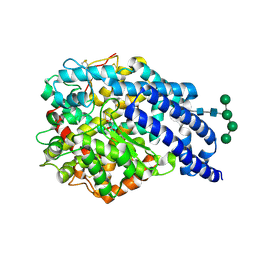 | | Crystal structure of AnCE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN CONVERTING ENZYME, CITRATE ANION, ... | | Authors: | Akif, M, Georgiadis, D, Mahajan, A, Dive, V, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Resolution Crystal Structures of Drosophila Melanogaster Angiotensin Converting Enzyme in Complex with Novel Inhibitors and Anti- Hypertensive Drugs.
J.Mol.Biol., 400, 2010
|
|
2WR0
 
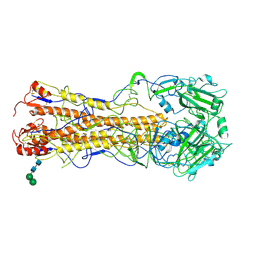 | | Structures of influenza H2 Hemagglutinins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, ... | | Authors: | Liu, J, Stevens, D, Haire, L, Coombs, P, Russell, R, Gamblin, S, Skehel, J. | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | From the Cover: Structures of Receptor Complexes Formed by Hemagglutinins from the Asian Influenza Pandemic of 1957.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4RHN
 
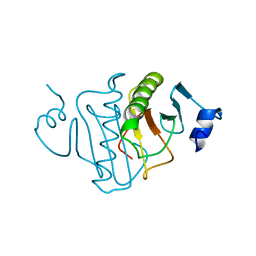 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH ADENOSINE | | Descriptor: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN, alpha-D-ribofuranose | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-26 | | Release date: | 1997-06-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
4Y7P
 
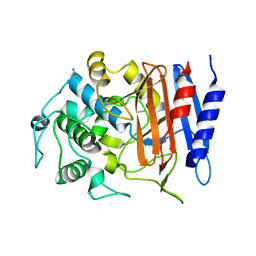 | | Structure of alkaline D-peptidase from Bacillus cereus | | Descriptor: | Alkaline D-peptidase, THIOCYANATE ION | | Authors: | Nakano, S, Okazaki, S, Ishitsubo, E, Kawahara, N, Komeda, H, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-02-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational analysis of peptide recognition mechanism of class-C type penicillin binding protein, alkaline D-peptidase from Bacillus cereus DF4-B
Sci Rep, 5, 2015
|
|
7LS0
 
 | | Structure of the Human ALK GRD bound to AUG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALK tyrosine kinase receptor fused with ALK and LTK ligand 2, CITRIC ACID | | Authors: | Stayrook, S, Li, T, Klein, D.E. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
7LI9
 
 | |
7LIA
 
 | |
7LI8
 
 | |
5DFM
 
 | | Structure of Tetrahymena telomerase p19 fused to MBP | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein,Telomerase-associated protein 19, SULFATE ION, ... | | Authors: | Chan, H, Cascio, D, Sawaya, M.R, Feigon, J. | | Deposit date: | 2015-08-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions.
Science, 350, 2015
|
|
7MPG
 
 | |
