8UIR
 
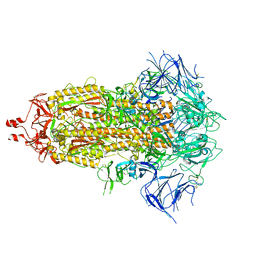 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer consensus (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
7Y8X
 
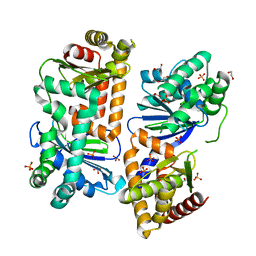 | | Crystal structure of AlbEF homolog from Quasibacillus thermotolerans in complex with Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, AlbE homolog, AlbF homolog, ... | | Authors: | Ishida, K, Nakamura, A, Kojima, S. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the AlbEF complex involved in subtilosin A biosynthesis.
Structure, 30, 2022
|
|
8V0S
 
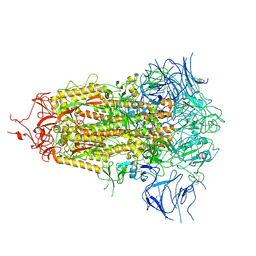 | | SARS-CoV-2 Omicron-XBB.1.5 3-RBD down Spike Protein Trimer 2 (S-GSAS-Omicron-XBB.1.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
6R7E
 
 | | N-terminally reversed variant of FimA E. coli with alanine insertion at position 20 | | Descriptor: | FimA, SULFATE ION | | Authors: | Zyla, D, Echeverria, B, Glockshuber, R. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Donor strand sequence, rather than donor strand orientation, determines the stability and non-equilibrium folding of the type 1 pilus subunit FimA.
J.Biol.Chem., 295, 2020
|
|
5N8V
 
 | | Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. | | Descriptor: | 1-(2-azanylethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Dawidowski, M, Emmanouilidis, L, Sattler, M, Popowicz, G.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.
Science, 355, 2017
|
|
5WQO
 
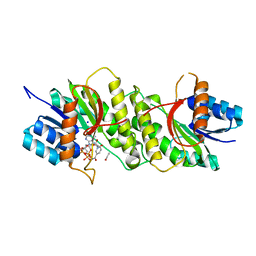 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition I) | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable dehydrogenase, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
6DNS
 
 | | Endo-fucoidan hydrolase MfFcnA9 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
6YUS
 
 | | Capsule O-acetyltransferase of Neisseria meningitidis serogroup A H228A mutant in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cramer, J.T, Fiebig, T, Fedorov, R, Muehlenhoff, M. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic basis of capsule O-acetylation in Neisseria meningitidis serogroup A.
Nat Commun, 11, 2020
|
|
5NDD
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ8838 at 2.8 angstrom resolution | | Descriptor: | (~{S})-(4-fluoranyl-2-propyl-phenyl)-(1~{H}-imidazol-2-yl)methanol, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, PHOSPHATE ION, ... | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
4RYZ
 
 | |
5HQM
 
 | | Structure function studies of R. palustris RubisCO (R. palustris/R. rubrum chimera) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Arbing, M.A, Shin, A, Cascio, D, Satagopan, S, Tabita, F.R. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure function studies of R. palustris RubisCO.
To Be Published
|
|
8V0R
 
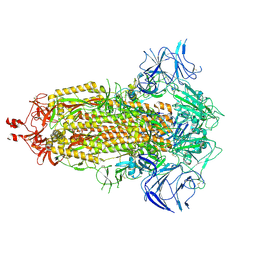 | | SARS-CoV-2 Omicron-XBB.1.5 3-RBD down Spike Protein Trimer 1 (S-GSAS-Omicron-XBB.1.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
5DKK
 
 | |
6PGZ
 
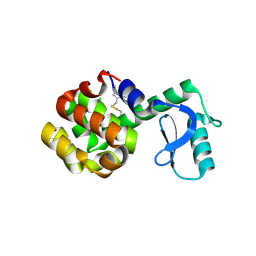 | | MTSL labelled T4 lysozyme pseudo-wild type V75C mutant | | Descriptor: | CHLORIDE ION, Endolysin, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
5HIQ
 
 | |
7OVJ
 
 | | Protein kinase MKK7 in complex with difluoro-phenethyltriazole-substituted pyrazolopyrimidine | | Descriptor: | 1-[(3~{R})-3-[4-azanyl-3-[1-[2,2-bis(fluoranyl)-2-phenyl-ethyl]-1,2,3-triazol-4-yl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
9E15
 
 | | Alpha-Delta heterodimeric form of soluble hydrogenase I from Pyrococcus furiosus. Data processed and model refined in P1 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Lanzilotta, W.N, McTernan, P.M, Adams, M.W.W. | | Deposit date: | 2024-10-21 | | Release date: | 2025-07-16 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the biotechnologically relevant reversible NADPH-oxidizing NiFe-hydrogenase from P.furiosus.
Structure, 33, 2025
|
|
6DFW
 
 | | TCR 8F10 in complex with IAg7-p8G9E | | Descriptor: | 8F10 alpha chain, 8F10 beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
7OVK
 
 | | Protein kinase MKK7 in complex with 5-bromo-2-hydroxyphenyl-substituted pyrazolopyrimidine | | Descriptor: | 1-[(3~{R})-3-[4-azanyl-3-[1-(5-bromanyl-2-oxidanyl-phenyl)-1,2,3-triazol-4-yl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, Dual specificity mitogen-activated protein kinase kinase 7, GLYCEROL | | Authors: | Kleinboelting, S, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Optimization of Covalent MKK7 Inhibitors via Crude Nanomole-Scale Libraries.
J.Med.Chem., 65, 2022
|
|
7F09
 
 | | Crystal structure of the HLH-Lz domain of human TFE3 | | Descriptor: | 1,2-ETHANEDIOL, Transcription factor E3, ZINC ION | | Authors: | Yang, G, Li, P, Liu, Z, Wu, S, Zhuang, C, Qiao, H, Fang, P, Wang, J. | | Deposit date: | 2021-06-03 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the dimerization mechanism of human transcription factor E3.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
4Z9P
 
 | | Crystal structure of Ebola virus nucleoprotein core domain at 1.8A resolution | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Dong, S.S, Yang, P, Li, G.B, Liu, B.C, Yang, C, Rao, Z.H. | | Deposit date: | 2015-04-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution.
Protein Cell, 6, 2015
|
|
7ALE
 
 | | Crystal structure of human PAICS in complex with inhibitor 69 | | Descriptor: | (2~{S})-2-[[5-azanyl-1-[(2~{R},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]imidazol-4-yl]car bonylamino]butanedioic acid, 2-azanyl-~{N}-[2-bromanyl-5-[4-[3-(dimethylamino)propylsulfonyl]piperazin-1-yl]phenyl]-1,3-oxazole-4-carboxamide, Multifunctional protein ADE2 | | Authors: | Skerlova, J, Marttila, P, Unterlass, J, Jemth, A.-S, Henriksson, M, Wakchaure, P, Grube, M, Warpman Berglund, U, Homan, E, Helleday, T, Stenmark, P. | | Deposit date: | 2020-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Cellular and biochemical validation of a potent PAICS inhibitor
To Be Published
|
|
6OCZ
 
 | | Crystal Structure of Mycobacterium tuberculosis Proteasome in Complex with Phenylimidazole-based Inhibitor A86 | | Descriptor: | CITRIC ACID, DIMETHYLFORMAMIDE, N-{(2S)-1-({(2S)-1-[(2,4-difluorobenzyl)amino]-1-oxopropan-2-yl}amino)-1,4-dioxo-4-[(2R)-2-phenylpyrrolidin-1-yl]butan-2-yl}-5-methyl-1,2-oxazole-3-carboxamide (non-preferred name), ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2019-03-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Selective Phenylimidazole-Based Inhibitors of theMycobacterium tuberculosisProteasome.
J.Med.Chem., 62, 2019
|
|
8V4J
 
 | | Phosphoheptose isomerase GMHA from Burkholderia pseudomallei bound to inhibitor Mut148233 | | Descriptor: | 1-deoxy-1-[formyl(hydroxy)amino]-5-O-phosphono-D-ribitol, CHLORIDE ION, Phosphoheptose isomerase, ... | | Authors: | Junop, M.S, Brown, C, Szabla, R. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Potentiating Activity of GmhA Inhibitors on Gram-Negative Bacteria.
J.Med.Chem., 67, 2024
|
|
8JYZ
 
 | | Cryo-EM structure of RCD-1 pore from Neurospora crassa | | Descriptor: | Gasdermin-like protein rcd-1-1, Gasdermin-like protein rcd-1-2 | | Authors: | Hou, Y.J, Sun, Q, Li, Y, Ding, J. | | Deposit date: | 2023-07-04 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cleavage-independent activation of ancient eukaryotic gasdermins and structural mechanisms.
Science, 384, 2024
|
|
