8F10
 
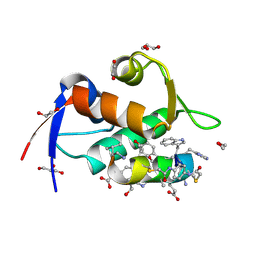 | | Structure of the MDM2 P53 binding domain in complex with H102, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
6GX2
 
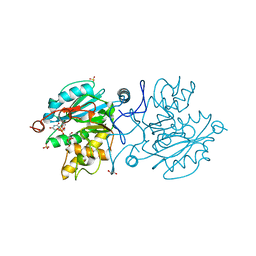 | |
6QE9
 
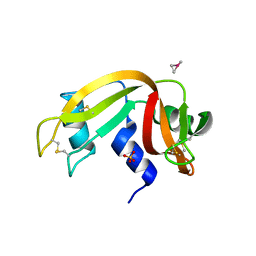 | | The X-ray structure of the adduct formed in the reaction between bovine pancreatic ribonuclease and complex I, a pentacoordinate Pt(II) compound containing 2,9-dimethyl-1,10-phenanthroline, dimethylfumarate, methyl and iodine as ligands | | Descriptor: | Ribonuclease pancreatic, SULFATE ION, pentacoordinate Pt(II) compound | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2019-01-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Reaction with Proteins of a Five-Coordinate Platinum(II) Compound.
Int J Mol Sci, 20, 2019
|
|
9H6X
 
 | | Cryo-EM structure of lipid-bound human myoferlin (25 mol% DOPS/5 mol% PI(4,5)P2 nanodisc) | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, CALCIUM ION, Myoferlin | | Authors: | Cretu, C, Moser, T, Preobraschenski, J. | | Deposit date: | 2024-10-25 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural insights into lipid membrane binding by human ferlins.
Embo J., 44, 2025
|
|
6BP1
 
 | |
8F16
 
 | | Structure of the STUB1 TPR domain in complex with H203, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
5HMI
 
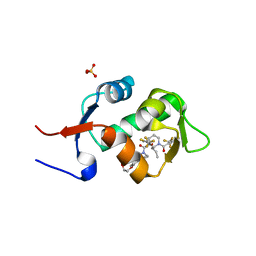 | | HDM2 in complex with a 3,3-Disubstituted Piperidine | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, {4-[2-(2-hydroxyethoxy)phenyl]piperazin-1-yl}[(2R,3S)-2-propyl-1-{[4-(trifluoromethyl)pyridin-3-yl]carbonyl}-3-{[5-(trifluoromethyl)thiophen-3-yl]oxy}piperidin-3-yl]methanone | | Authors: | Scapin, G. | | Deposit date: | 2016-01-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Novel 3,3-Disubstituted Piperidines as Orally Bioavailable, Potent, and Efficacious HDM2-p53 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5J6N
 
 | | Crystal Structure of Hsp90-alpha N-domain L107A mutant in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propyl-benzenesulfonamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propylbenzene-1-sulfonamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
5M6B
 
 | |
6HX7
 
 | | Crystal structure of human R180T variant of ORNITHINE AMINOTRANSFERASE at 1.8 Angstrom | | Descriptor: | Ornithine aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Giardina, G, Montioli, R, Cellini, B, Cutruzzola, F, Borri Voltattorni, C. | | Deposit date: | 2018-10-16 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | R180T variant of delta-ornithine aminotransferase associated with gyrate atrophy: biochemical, computational, X-ray and NMR studies provide insight into its catalytic features.
Febs J., 286, 2019
|
|
8BW1
 
 | | Yeast 20S proteasome in complex with an engineered fellutamide derivative (C14QAL) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bozhueyuek, K.A.J, Praeve, L, Kegler, C, Kaiser, S, Shi, Y, Kuttenlochner, W, Schenk, L, Groll, M, Hochberg, G.K.A, Bode, H.B. | | Deposit date: | 2022-12-06 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Evolution-inspired engineering of nonribosomal peptide synthetases.
Science, 383, 2024
|
|
5M75
 
 | |
9HH5
 
 | | Crystal Structure of nsp15 Endoribonuclease from SARS CoV-2 in Complex with Sepantronium (YM-155) | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nsp15 - Uridylate-specific endoribonuclease, ... | | Authors: | Chatziefthymiou, S.D, Kolbe, M, Hakanpaeae, J, Labahn, J, Windshuegel, B. | | Deposit date: | 2024-11-21 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Identification, validation, and characterization of approved and investigational drugs interfering with the SARS-CoV-2 endoribonuclease Nsp15.
Protein Sci., 34, 2025
|
|
5J82
 
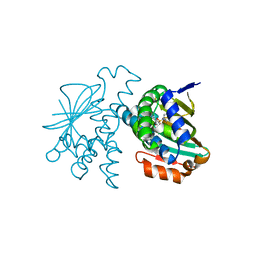 | | Crystal Structure of Hsp90-alpha N-domain in complex 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-isopropyl-N-methyl-benzenesulfonamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methyl-N-(propan-2-yl)benzene-1-sulfonamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
8EXU
 
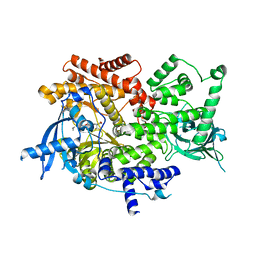 | | Crystal structure of PI3K-alpha in complex with compound 30 | | Descriptor: | (2S)-2-cyclopropyl-2-({(4S,11aM)-2-[(4S)-2-oxo-4-(trifluoromethyl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}amino)acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
5DC8
 
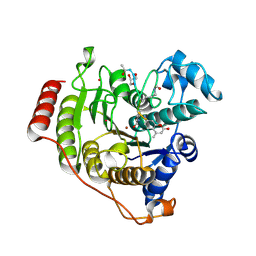 | | Crystal structure of H142A-Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | Fluor-de-Lys tetrapeptide assay substrate, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Lee, M.S, Christianson, D.W. | | Deposit date: | 2015-08-23 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | General Base-General Acid Catalysis in Human Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
5LT4
 
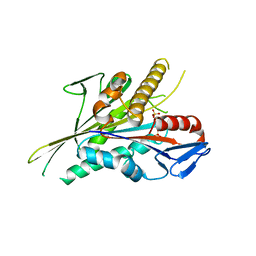 | |
6ZAA
 
 | | PI3K Delta in complex with methoxy(methylsulfamoyl)pyridinylN(methylpiperidinyl)dihydrobenzoxazinecarboxamide | | Descriptor: | 4-[6-methoxy-5-(methylsulfamoyl)pyridin-3-yl]-~{N}-(1-methylpiperidin-4-yl)-2,3-dihydro-1,4-benzoxazine-6-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Hardy, C.J, Spencer, J.A, Rowland, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Design and Development of a Macrocyclic Series Targeting Phosphoinositide 3-Kinase delta.
Acs Med.Chem.Lett., 11, 2020
|
|
4RYX
 
 | | Crystal structure of RPE65 in complex with emixustat and palmitate, P6522 crystal form | | Descriptor: | (1R)-3-amino-1-[3-(cyclohexylmethoxy)phenyl]propan-1-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, FE (II) ION, ... | | Authors: | Kiser, P.D, Palczewski, K. | | Deposit date: | 2014-12-17 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular pharmacodynamics of emixustat in protection against retinal degeneration.
J.Clin.Invest., 125, 2015
|
|
8FX2
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-HP | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8WYH
 
 | |
6FJ4
 
 | |
6Z7M
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 (3R,4R)-N-cyclohexyl-4-((3-methyl-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl)amino)piperidine-3-carboxamide | | Descriptor: | (3R,4R)-N-cyclohexyl-4-((3-methyl-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl)amino)piperidine-3-carboxamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2020-05-31 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | GSK789: A Selective Inhibitor of the First Bromodomains (BD1) of the Bromo and Extra Terminal Domain (BET) Proteins.
J.Med.Chem., 63, 2020
|
|
9HQJ
 
 | | UP1 in complex with Z86417414 | | Descriptor: | 2-fluoranyl-~{N}-(1,3,4-thiadiazol-2-yl)benzamide, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-12-16 | | Release date: | 2024-12-25 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNPA1 via cryptic pockets mapping coupled with X-ray fragment screening.
J.Biol.Chem., 301, 2025
|
|
9F4V
 
 | | UP1 in complex with EN300-118084 | | Descriptor: | 3-(difluoromethyl)-1-methyl-1H-pyrazole-4-carboxamide, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNPA1 via cryptic pockets mapping coupled with X-ray fragment screening.
J.Biol.Chem., 301, 2025
|
|
