7XS6
 
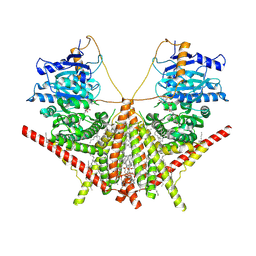 | | structure of a membrane-integrated glycosyltransferase with inhibitor | | Descriptor: | (19R,22S)-25-amino-22-hydroxy-22-oxido-16-oxo-17,21,23-trioxa-22lambda~5~-phosphapentacosan-19-yl (9Z)-hexadec-9-enoate, (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase 1, ... | | Authors: | Wu, Y.N, Zhang, M, Yang, Y.Z, Ding, X.Y, Liu, X.T, Zhang, M.J, Yu, H.J. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and mechanism of chitin synthase and its inhibition by antifungal drug Nikkomycin Z.
Cell Discov, 8, 2022
|
|
8Z00
 
 | | Tribolium castaneum ABCH-9C in complex with LMNG and ceramide(d36:1) | | Descriptor: | (~{E})-~{N}-[(2~{S},3~{R})-1,3-bis(oxidanyl)octadecan-2-yl]octadec-9-enamide, ABC transporter G family member 23-like Protein, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Yang, Q, Chen, J, Zhou, Y. | | Deposit date: | 2024-04-08 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of ceramide transportation and its regulation by ABCH transporter
To Be Published
|
|
8Z01
 
 | | Tribolium castaneum ABCH-9C(E224Q) mutant in complex with LMNG and ceramide(d36:1) | | Descriptor: | (~{E})-~{N}-[(2~{S},3~{R})-1,3-bis(oxidanyl)octadecan-2-yl]octadec-9-enamide, ABC transporter G family member 23-like Protein, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Yang, Q, Chen, J, Zhou, Y. | | Deposit date: | 2024-04-08 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of ceramide transportation and its regulation by ABCH transporter
To Be Published
|
|
9DVN
 
 | | Structure of the phosphate exporter XPR1/SLC53A1, InsP8-bound, intracellular gate closed state | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Zhu, Q, Diver, M.M. | | Deposit date: | 2024-10-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Transport and InsP 8 gating mechanisms of the human inorganic phosphate exporter XPR1.
Nat Commun, 16, 2025
|
|
9G0C
 
 | |
4NSM
 
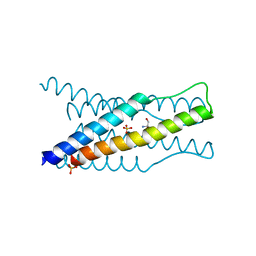 | | crystal structure of the streptococcal collagen-like protein 2 globular domain from invasive M3-type group A Streptococcus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Collagen-like protein SclB, SULFATE ION | | Authors: | Berisio, R, Squeglia, F, Lukomski, S, Bachert, B. | | Deposit date: | 2013-11-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Streptococcal Collagen-like Protein 2 Globular Domain from Invasive M3-type Group A Streptococcus Shows Significant Similarity to Immunomodulatory HIV Protein gp41.
J.Biol.Chem., 289, 2014
|
|
9DVP
 
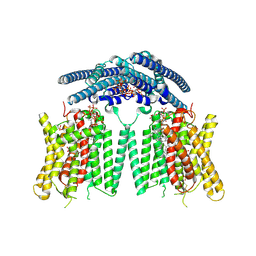 | | Structure of the phosphate exporter XPR1/SLC53A1, Pi and InsP8-bound, intracellular gate closed state | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Zhu, Q, Diver, M.M. | | Deposit date: | 2024-10-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Transport and InsP 8 gating mechanisms of the human inorganic phosphate exporter XPR1.
Nat Commun, 16, 2025
|
|
9DVM
 
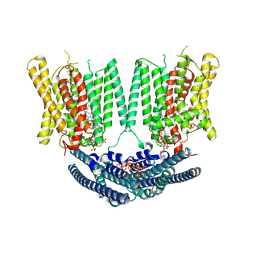 | | Structure of the phosphate exporter XPR1/SLC53A1, InsP8-bound, intracellular gate open/intracellular gate closed state | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Zhu, Q, Diver, M.M. | | Deposit date: | 2024-10-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Transport and InsP 8 gating mechanisms of the human inorganic phosphate exporter XPR1.
Nat Commun, 16, 2025
|
|
9DVO
 
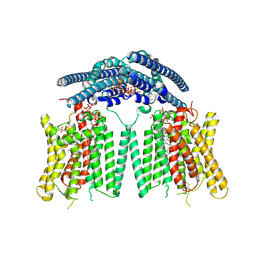 | | Structure of the phosphate exporter XPR1/SLC53A1, Pi and InsP8-bound, intracellular gate open/intracellular gate closed state | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Zhu, Q, Diver, M.M. | | Deposit date: | 2024-10-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transport and InsP 8 gating mechanisms of the human inorganic phosphate exporter XPR1.
Nat Commun, 16, 2025
|
|
7YIS
 
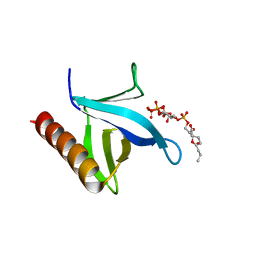 | | Crystal structure of N-terminal PH domain of ARAP3 protein in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Zhang, Y.J, Liu, Y.R, Wu, B. | | Deposit date: | 2022-07-18 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insights Uncover the Specific Phosphoinositide Recognition by the PH1 Domain of Arap3.
Int J Mol Sci, 24, 2023
|
|
8DGT
 
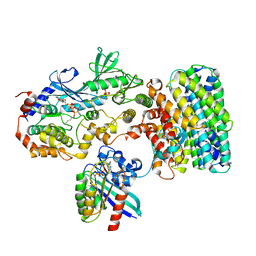 | | Cryo-EM structure of a RAS/RAF complex (state 2) | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Eck, M.J, Jeon, H, Park, E, Rawson, S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a RAS/RAF recruitment complex.
Nat Commun, 14, 2023
|
|
4QND
 
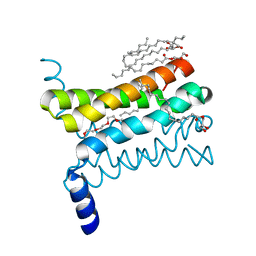 | | Crystal structure of a SemiSWEET | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Chemical transport protein, ... | | Authors: | Yan, X, Yuyong, T, Liang, F, Perry, K. | | Deposit date: | 2014-06-17 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structures of bacterial homologues of SWEET transporters in two distinct conformations.
Nature, 515, 2014
|
|
8R3A
 
 | | NT-26 Arsenite oxidase B F108C-G123C | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Engrola, F, Santos-Silva, T, Correia, M.A.S, Romao, M.J. | | Deposit date: | 2023-11-08 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | NT-26 Arsenite oxidase B F108C-G123C
To Be Published
|
|
4IYK
 
 | |
6YEN
 
 | | Crystal structure of AmpC from E. coli with Taniborbactam (VNRX-5133) | | Descriptor: | (10aR)-2-(((1r,4R)-4-((2-aminoethyl)amino)cyclohexyl)methyl)-6-carboxy-4-hydroxy-4,10a-dihydro-10H-benzo[5,6][1,2]oxaborinino[2,3-b][1,4,2]oxazaborol-4-uide, (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, 1,2-ETHANEDIOL, ... | | Authors: | Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Bicyclic Boronates as Potent Inhibitors of AmpC, the Class C beta-Lactamase from Escherichia coli .
Biomolecules, 10, 2020
|
|
4QP0
 
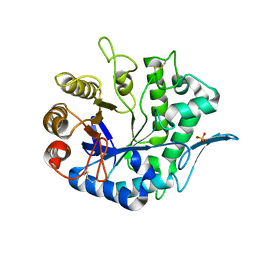 | | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei | | Descriptor: | Endo-beta-mannanase, SULFATE ION | | Authors: | Zheng, Q.J, Peng, Z, Liu, Y, Yan, Q.J, Chen, Z.Z, Qin, Z. | | Deposit date: | 2014-06-22 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6EIX
 
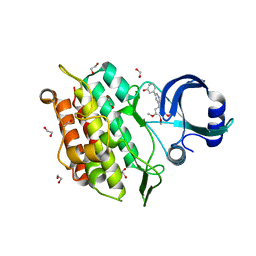 | | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with a 2-aminopyridine inhibitor K02288 | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1 | | Authors: | Williams, E.P, Canning, P, Sanvitale, C.E, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2017-09-19 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with K02288
To be published
|
|
4QNS
 
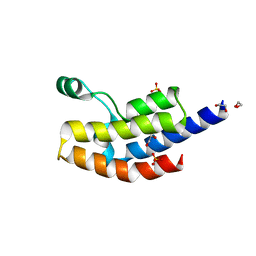 | | Crystal structure of bromodomain from Plasmodium faciparum GCN5, PF3D7_0823300 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone acetyltransferase GCN5, ... | | Authors: | Fonseca, M, Tallant, C, Knapp, S, Loppnau, P, von Delft, F, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Crystal structure of bromodomain from Plasmodium faciparum GCN5, PF3D7_0823300
TO BE PUBLISHED
|
|
7AXJ
 
 | | Crystal structure of the hPXR-LBD in complex with estradiol and clotrimazole | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, ESTRADIOL, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Delfosse, V, Granell, M, Blanc, P, Bourguet, W. | | Deposit date: | 2020-11-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insights into the synergistic activation of the RXR-PXR heterodimer by endocrine disruptor mixtures.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6GKV
 
 | | Crystal structure of Coclaurine N-Methyltransferase (CNMT) bound to N-methylheliamine and SAH | | Descriptor: | 6,7-dimethoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-2-ium, Coclaurine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Dunstan, M.S, Levy, C.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Biocatalytic Scope of Coclaurine N-Methyltransferase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8GPW
 
 | | Structure of Penicillin-binding protein 3 (PBP3) from Klebsiella pneumoniae with ligand 18G | | Descriptor: | 1-[(~{Z})-[1-(2-azanyl-1,3-thiazol-4-yl)-2-[[(2~{S})-3-methyl-1-oxidanylidene-3-(sulfooxyamino)butan-2-yl]amino]-2-oxidanylidene-ethylidene]amino]oxycyclopropane-1-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Song, D.Q, Li, Y.H. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of Penicillin-binding protein 3 (PBP3) from Klebsiella pneumoniae with ligand 18G at 2.06 Angstroms resolution
To Be Published
|
|
9FWH
 
 | | Crystal Structure of SARS-CoV-2 NSP10-ExoN in complex with VT00019 | | Descriptor: | (4R)-4-phenyl-1,2-thiazolidine 1,1-dioxide, Guanine-N7 methyltransferase nsp14, Non-structural protein 10, ... | | Authors: | Krojer, T, Kozielski, F, Sele, C, Nyblom, M, Fisher, S.Z, Knecht, W. | | Deposit date: | 2024-06-30 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of SARS-CoV-2 NSP10-ExoN in complex with VT00019
To Be Published
|
|
6A20
 
 | | Crystal Structure of auto-inhibited Kinesin-3 KIF13B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEXAETHYLENE GLYCOL, Kinesin family member 13B, ... | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-06-08 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Coiled-coil 1-mediated fastening of the neck and motor domains for kinesin-3 autoinhibition.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8RZA
 
 | | Ribonuclease W | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Probable ribonuclease FAU-1, ... | | Authors: | Vayssieres, M, Blaud, M, Leulliot, N. | | Deposit date: | 2024-02-12 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | RNase W, a conserved ribonuclease family with a novel active site.
Nucleic Acids Res., 52, 2024
|
|
9G1K
 
 | | Fragment screening of FosAKP, cryo structure in complex with fragment F2X-entry A09 | | Descriptor: | 1,2-ETHANEDIOL, 2-sulfanylpyridine-3-carboximidamide, Fosfomycin resistance protein, ... | | Authors: | Guenther, S, Galchenkova, M, Fischer, P, Reinke, P.Y.A, Falke, S, Thekku Veedu, S, Rodrigues, A.C, Senst, J, Meents, A. | | Deposit date: | 2024-07-10 | | Release date: | 2025-07-23 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Room-temperature X-ray fragment screening with serial crystallography.
Nat Commun, 16, 2025
|
|
