7X2T
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (CVB1-M:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
8EI7
 
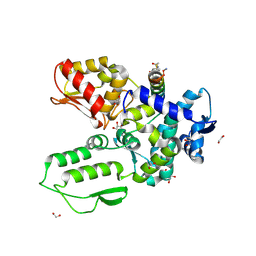 | | Crystal structure of the WWP2 HECT domain in complex with H304, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Li, K, Tokareva, O.S, Thomson, T.M, Verdine, G.L, McGee, J.H. | | Deposit date: | 2022-09-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Recognition and reprogramming of E3 ubiquitin ligase surfaces by alpha-helical peptides.
Nat Commun, 14, 2023
|
|
7X37
 
 | | Cryo-EM structure of Coxsackievirus B1 A particle in complex with nAb 2E6 (CVB1-A:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3C
 
 | | Cryo-EM structure of Coxsackievirus B1 muture virion in complex with nAbs 8A10 and 5F5 (CVB1-M:8A10:5F5) | | Descriptor: | 5F5 heavy chain, 5F5 light chain, 8A10 heavy chain, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7V46
 
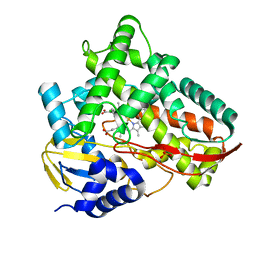 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with ortho-chlorotoluene. | | Descriptor: | 1-chloranyl-3-methyl-benzene, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7X3E
 
 | | Cryo-EM structure of Coxsackievirus B1 pre-A-particle in complex with nAb 9A3 (CVB1-pre-A:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
8F9T
 
 | |
9F39
 
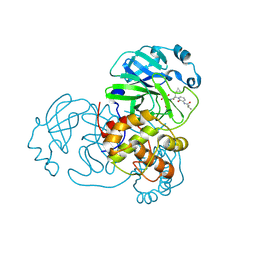 | | Crystal structure of SARS-CoV-2 Mpro in complex with RK-54 | | Descriptor: | (2R,3R)-3-[[(2S)-3-cyclopropyl-2-[3-(2-methylpropanoylamino)-2-oxidanylidene-pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5 | | Authors: | El kilani, H, Hilgenfeld, R. | | Deposit date: | 2024-04-25 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Optimization of Pyridone alpha-Ketoamides as Inhibitors of the SARS-CoV-2 Main Protease.
J.Med.Chem., 68, 2025
|
|
7X3D
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 9A3 (CVB1-M:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X40
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-28 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
9J52
 
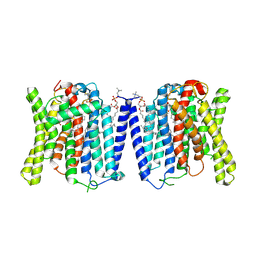 | | CryoEM structure of human XPR1 in complex with phosphate in state B | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PHOSPHATE ION, Solute carrier family 53 member 1 | | Authors: | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | Deposit date: | 2024-08-11 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
8F9U
 
 | |
7X46
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X47
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4K
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7UEL
 
 | |
9J51
 
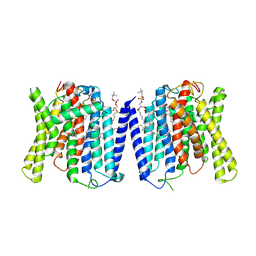 | | CryoEM structure of human XPR1 in complex with phosphate in state A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PHOSPHATE ION, Solute carrier family 53 member 1 | | Authors: | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | Deposit date: | 2024-08-11 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
9J53
 
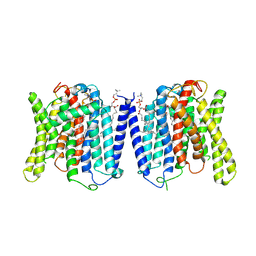 | | CryoEM structure of human XPR1 in complex with phosphate in state C | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PHOSPHATE ION, Solute carrier family 53 member 1 | | Authors: | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | Deposit date: | 2024-08-11 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
6IH7
 
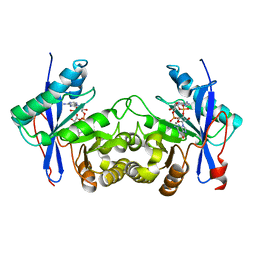 | | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - 3',3'-cGAMP bound form | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, cyclic di nucleotide phoshodiesterase | | Authors: | Yadav, M, Pal, K, Sen, U. | | Deposit date: | 2018-09-28 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of c-di-GMP/cGAMP degrading phosphodiesterase VcEAL: identification of a novel conformational switch and its implication.
Biochem.J., 476, 2019
|
|
6AQ3
 
 | |
7X4M
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10, 2E6 and 9A3) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
9J7H
 
 | | Crystal structure of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (DAHP synthase) from Providencia alcalifaciens complexed with quinic acid | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Phospho-2-dehydro-3-deoxyheptonate aldolase | | Authors: | Jangid, K, Mahto, J.K, Kumar, K.A, Kumar, P. | | Deposit date: | 2024-08-19 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and biochemical analyses reveal quinic acid inhibits DAHP synthase a key player in shikimate pathway.
Arch.Biochem.Biophys., 763, 2025
|
|
9F8A
 
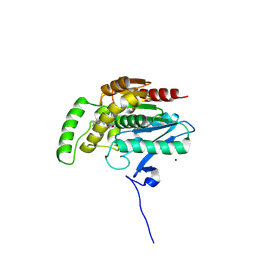 | | Crystal structure of human monoacylglycerol lipase in complex with compound 7a | | Descriptor: | 6-[4-(phenylmethyl)piperidin-1-yl]carbonyl-4~{H}-1,4-benzoxazin-3-one, Monoglyceride lipase, SODIUM ION | | Authors: | Kuhn, B, Ritter, M, Hornsperger, B, Bell, C, Kocer, B, Rombach, D, Richter, H, Grether, U, Gobbi, L, Kuratli, M, Collin, L, Benz, J. | | Deposit date: | 2024-05-06 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure-Guided Discovery of cis -Hexahydro-pyrido-oxazinones as Reversible, Drug-like Monoacylglycerol Lipase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8F9F
 
 | |
5L2V
 
 | | Catalytic domain of LPMO Lmo2467 from Listeria monocytogenes | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, COPPER (II) ION, Chitin-binding protein | | Authors: | Light, S.H, Agostoni, M, Marletta, M.A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-02 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalytic domain of LPMO Lmo2467 from Listeria monocytogenes
To Be Published
|
|
