7BJ3
 
 | | ScpA from Streptococcus pyogenes, S512A active site mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Kagawa, T.F, O'Connell, M.R, Cooney, J.C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease.
Comput Struct Biotechnol J, 19, 2021
|
|
3K8K
 
 | | Crystal structure of SusG | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Alpha-amylase, ... | | Authors: | Koropatkin, N.M, Smith, T.J. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SusG: A Unique Cell-Membrane-Associated alpha-Amylase from a Prominent Human Gut Symbiont Targets Complex Starch Molecules.
Structure, 18, 2010
|
|
8HUM
 
 | | X-ray structure of human PPAR gamma ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
5HLP
 
 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH BRD3937 | | Descriptor: | 4-(2-methoxyphenyl)-3,7,7-trimethyl-1,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
6ZBB
 
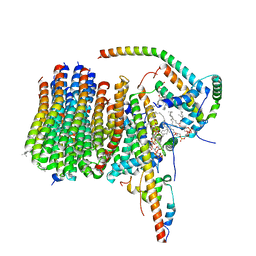 | | bovine ATP synthase Fo domain | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7SYQ
 
 | | Structure of the wt IRES and 40S ribosome ternary complex, open conformation. Structure 11(wt) | | Descriptor: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYP
 
 | | Structure of the wt IRES and 40S ribosome binary complex, open conformation. Structure 10(wt) | | Descriptor: | 18S rRNA, HCV IRES, HCV IRES partially loaded mRNA portion, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
9FNN
 
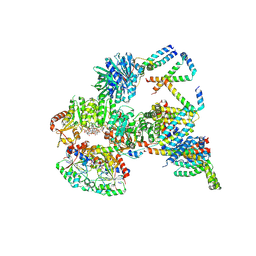 | | Cryo-EM structure of the c-di-GMP-saturated 'crown'less Bcs macrocomplex for cellulose secretion in E. coli | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ADENOSINE-5'-TRIPHOSPHATE, Cell division protein, ... | | Authors: | Anso, I, Krasteva, P.V. | | Deposit date: | 2024-06-10 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system.
Nat Commun, 15, 2024
|
|
7SYO
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head open. Structure 9(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
6RDN
 
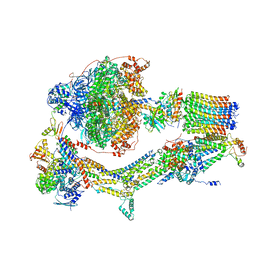 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1C, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
5KZN
 
 | | Metabotropic Glutamate Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
6RE3
 
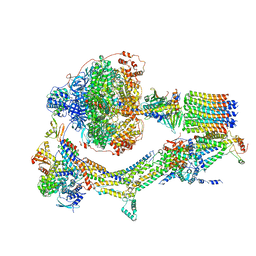 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2B, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
9FP0
 
 | | Cryo-EM structure of the 'crown'less Bcs macrocomplex for E. coli cellulose secretion in non-saturating c-di-GMP (local) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ADENOSINE-5'-TRIPHOSPHATE, Cell division protein, ... | | Authors: | Anso, I, Krasteva, P.V. | | Deposit date: | 2024-06-12 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system.
Nat Commun, 15, 2024
|
|
8ZRT
 
 | | Cryo-EM structure focused on the receptor of the ET-1 bound ETBR-DNGI complex | | Descriptor: | Endothelin receptor type B, Endothelin-1 | | Authors: | Tani, K, Maki-Yonekura, S, Kanno, R, Negami, T, Hamaguchi, T, Hall, M, Mizoguchi, A, Humbel, B.M, Terada, T, Yonekura, K, Doi, T. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structure of endothelin ET B receptor-G i complex in a conformation stabilized by unique NPxxL motif.
Commun Biol, 7, 2024
|
|
3QWN
 
 | |
6E84
 
 | | Crystal structure of the Corn aptamer in complex with TO | | Descriptor: | 1-methyl-4-[(Z)-(3-methyl-1,3-benzothiazol-2(3H)-ylidene)methyl]quinolin-1-ium, POTASSIUM ION, RNA (36-MER) | | Authors: | Sjekloca, L, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Binding between G Quadruplexes at the Homodimer Interface of the Corn RNA Aptamer Strongly Activates Thioflavin T Fluorescence.
Cell Chem Biol, 26, 2019
|
|
8YAJ
 
 | | ATAT-2 bound MEC-12/MEC-7 microtubule without acetyl-CoA | | Descriptor: | Alpha-tubulin N-acetyltransferase 2, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Lam, W.H, Yu, D, Zhai, Y, Ti, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-11-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Tubulin acetyltransferases access and modify the microtubule luminal K40 residue through anchors in taxane-binding pockets.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7T9N
 
 | | M22 Agonist Autoantibody bound to Human Thyrotropin receptor in complex with miniGs399 (composite structure) | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Faust, B, Cheng, Y, Manglik, A. | | Deposit date: | 2021-12-19 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Autoantibody mimicry of hormone action at the thyrotropin receptor.
Nature, 609, 2022
|
|
7QZ4
 
 | | BAZ2A bromodomain in complex with acetylpyrrole derivative compound 87 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-ethyl-5-[2-[4-[(1-ethylpiperidin-4-yl)methyl]piperazin-1-yl]-1,3-thiazol-4-yl]-2-methyl-1~{H}-pyrrol-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Cazzanelli, G, Caflisch, A, Lolli, G. | | Deposit date: | 2022-01-30 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a BAZ2A-Bromodomain Hit Compound by Fragment Growing.
Acs Med.Chem.Lett., 13, 2022
|
|
7BGW
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1011 | | Descriptor: | 14-3-3 protein sigma, 4-(2-phenylimidazol-1-yl)naphthalene-1-carbaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
6RBP
 
 | |
6Y4W
 
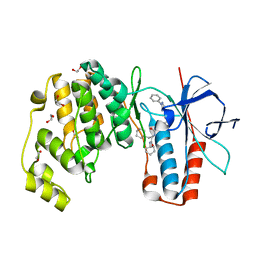 | | Crystal structure of p38 in complex with SR69 | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-(cyclohexylamino)-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Roehm, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Selective targeting of the alpha C and DFG-out pocket in p38 MAPK.
Eur.J.Med.Chem., 208, 2020
|
|
6YEB
 
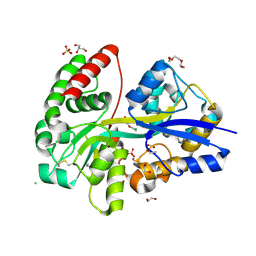 | | E.coli's Putrescine receptor PotF in its closed apo state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
6RDT
 
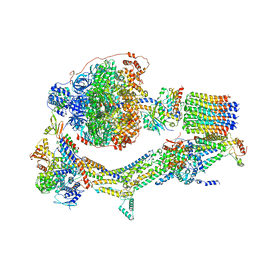 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1E, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
9EQ5
 
 | |
