5KGZ
 
 | | Phenol-soluble modulin Beta2 | | Descriptor: | Modulin Beta2 | | Authors: | Towle, K.M, Lohans, C.T, Acedo, J.Z, Van Belkum, M.J, Miskolzie, M, Vederas, J.C. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Phenol-Soluble Modulins alpha 1, alpha 3, and beta 2, Virulence Factors from Staphylococcus aureus.
Biochemistry, 55, 2016
|
|
5NA0
 
 | | TTK kinase domain in complex with a PEG-linked pyrimido-indolizine | | Descriptor: | Dual specificity protein kinase TTK, ~{N}-[3,5-diethyl-1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyrazol-4-yl]-2-[[2-methoxy-4-[4-(2-methoxyethanoyl)piperazin-1-yl]phenyl]amino]-5,6-dihydropyrimido[4,5-e]indolizine-7-carboxamide | | Authors: | Uitdehaag, J, Willemsen-Seegers, N, de Man, J, Buijsman, R.C, Zaman, G.J.R. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Target Residence Time-Guided Optimization on TTK Kinase Results in Inhibitors with Potent Anti-Proliferative Activity.
J. Mol. Biol., 429, 2017
|
|
5K5M
 
 | |
6GZV
 
 | | Identification of a druggable VP1-VP3 interprotomer pocket in the capsid of enteroviruses | | Descriptor: | 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]benzoic acid, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Geraets, J.A, Flatt, J.W, Domanska, A, Butcher, S.J. | | Deposit date: | 2018-07-05 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A novel druggable interprotomer pocket in the capsid of rhino- and enteroviruses.
Plos Biol., 17, 2019
|
|
8GGA
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #16 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGC
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #18 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGE
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #19 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
8GGF
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (transition intermediate #20 of 20) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
5K6I
 
 | | Crystal structure of prefusion-stabilized RSV F single-chain 9-10 DS-Cav1 A149C-Y458C, S46G-E92D-S215P-K465Q variant. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Joyce, M.G, Zhang, B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-24 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV.
Nat.Struct.Mol.Biol., 23, 2016
|
|
9KF5
 
 | | Crystal structure of Ferritin mutant(E53H/E57H) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Tian, J, Maity, B, Furuta, T, Pan, T, Abe, S, Ueno, T. | | Deposit date: | 2024-11-05 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Artificial Metal-Free Peroxidase Designed Using a Ferritin Cage for Bioinspired Catalysis.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9KF7
 
 | | Crystal structure of ferritin mutant (R52H/E56H/R59H/E63H) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Tian, J, Maity, B, Furuta, T, Pan, T, Abe, S, Ueno, T. | | Deposit date: | 2024-11-05 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Artificial Metal-Free Peroxidase Designed Using a Ferritin Cage for Bioinspired Catalysis.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9KF8
 
 | | Crystal structure of ferritin mutant (E53H/E57H/E60H/R64H) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Tian, J, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2024-11-05 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | An Artificial Metal-Free Peroxidase Designed Using a Ferritin Cage for Bioinspired Catalysis.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9KFA
 
 | | Crystal structure of ferritin mutantFr-H8 (R52H/E53H/E56H/E57H/R59H/E60H/E63H/R64H) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Tian, J, Maity, B, Furuta, T, Pan, T, Abe, S, Ueno, T. | | Deposit date: | 2024-11-05 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Artificial Metal-Free Peroxidase Designed Using a Ferritin Cage for Bioinspired Catalysis.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
170L
 
 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Zhang, X.-J, Weaver, L.H, Wozniak, A, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
8PSY
 
 | | ERK2 covelently bound to RU68 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{S},3~{R})-4-oxidanylidene-1-(phenylmethyl)cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
9LM3
 
 | | cryo-EM structure of retron Eco2 | | Descriptor: | DNA (5'-D(P*GP*GP*A)-3'), DNA (67-MER), RNA (5'-R(*GP*UP*GP*CP*CP*UP*GP*CP*AP*UP*GP*CP*GP*U)-3'), ... | | Authors: | Wang, Y.J, Wang, C, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2025-01-18 | | Release date: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of the RNA-mediated Retron-Eco2 oligomerization.
Cell Discov, 11, 2025
|
|
9KF9
 
 | | Crystal structure of ferritin mutant(E53H/E56H/E57H/R59H/E60H/E63H) | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Tian, J, Maity, B, Furuta, T, Pan, T, Ueno, T. | | Deposit date: | 2024-11-05 | | Release date: | 2025-09-24 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Artificial Metal-Free Peroxidase Designed Using a Ferritin Cage for Bioinspired Catalysis.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
7ATE
 
 | | Cytochrome c oxidase structure in P-state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Kolbe, F, Safarian, S, Michel, H. | | Deposit date: | 2020-10-30 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cytochrome c oxidase structure in P-state
To Be Published
|
|
8ZTB
 
 | |
8XOL
 
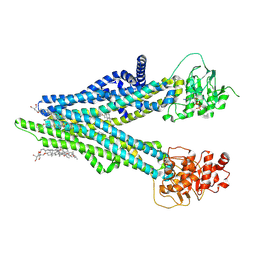 | | Cryo-EM structure of human ABCC4 with ANP bound in NBD1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
6Q38
 
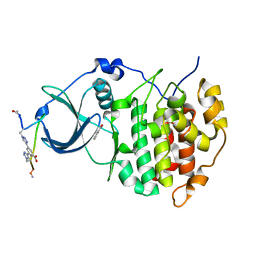 | | The Crystal structure of CK2a bound to P1-C4 | | Descriptor: | 3,5-bis(1-methyl-1,2,3-triazol-4-yl)benzoic acid, BENZOIC ACID, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Iegre, J, Baker, D, Tan, Y, Sore, H, Donovan, D, Spring, D, Chandra, V, Hyvonen, M. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Efficient development of stable and highly functionalised peptides targeting the CK2 alpha /CK2 beta protein-protein interaction.
Chem Sci, 10, 2019
|
|
8XOM
 
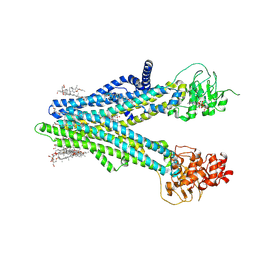 | | Cryo-EM structure of human ABCC4 in complex with ANP-bound in NBD1 and METHOTREXATE | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ATP-binding cassette sub-family C member 4, MAGNESIUM ION, ... | | Authors: | Zhang, P.F, Liu, Z. | | Deposit date: | 2024-01-01 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport.
Febs Lett., 598, 2024
|
|
5FGJ
 
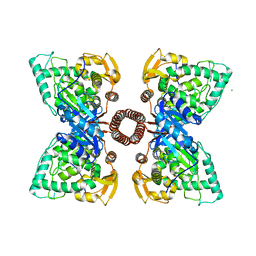 | | Structure of tetrameric rat phenylalanine hydroxylase, residues 1-453 | | Descriptor: | FE (III) ION, MAGNESIUM ION, Phenylalanine-4-hydroxylase | | Authors: | Taylor, A.B, Roberts, K.M, Fitzpatrick, P.F. | | Deposit date: | 2015-12-20 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Domain Movements upon Activation of Phenylalanine Hydroxylase Characterized by Crystallography and Chromatography-Coupled Small-Angle X-ray Scattering.
J.Am.Chem.Soc., 138, 2016
|
|
6YZ0
 
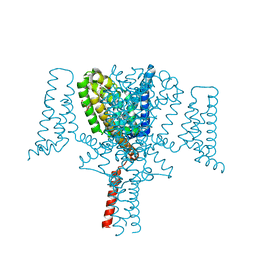 | | Full length Open-form Sodium Channel NavMs F208L in complex with Cannabidiol (CBD) | | Descriptor: | 2-[(1R,2R,5S)-5-methyl-2-(prop-1-en-2-yl)cyclohexyl]-5-pentylbenzene-1,3-diol, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Sait, L.G, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cannabidiol interactions with voltage-gated sodium channels.
Elife, 9, 2020
|
|
6QPB
 
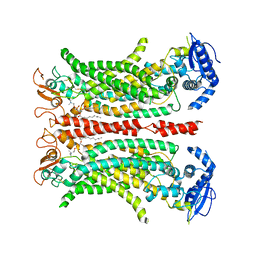 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
