8D7S
 
 | |
8YTK
 
 | | Crystal structure of human prolyl-tRNA synthetase in complex with inhibitor | | Descriptor: | (2~{S})-~{N}-[3-(4-azanylquinazolin-7-yl)phenyl]sulfonylpyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, Bifunctional glutamate/proline--tRNA ligase | | Authors: | Luo, Z, Zhou, H. | | Deposit date: | 2024-03-26 | | Release date: | 2025-04-02 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Development of potent inhibitors targeting bacterial prolyl-tRNA synthetase through fluorine scanning-directed activity tuning.
Eur.J.Med.Chem., 291, 2025
|
|
7OWB
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with a region from 10-hydroxylase CalMB | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,5,7-trihydroxy-6,11-dioxo-4-{[2,3,6-trideoxy-3-(dimethylamino)-alpha-L-lyxo-hexopyranosyl]oxy}-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
8D8B
 
 | |
7RYX
 
 | | S. CEREVISIAE CYP51 COMPLEXED WITH VT-1129 | | Descriptor: | (2R)-2-(2,4-difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-{5-[4-(trifluoromethoxy)phenyl]pyridin-2-yl}propan-2-ol, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ruma, Y.N, Sagatova, A, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterisation of Candida parapsilosis CYP51 as a Drug Target Using Saccharomyces cerevisiae as Host.
J Fungi, 8, 2022
|
|
8TLX
 
 | |
8INY
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) K90R Mutant in Complex with Inhibitor ensitrelvir | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
8INX
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) G15S Mutant in Complex with Inhibitor ensitrelvir | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants.
Commun Biol, 6, 2023
|
|
8P1O
 
 | | Solubilizer tag effect on PD-L1/inhibitor binding properties for m-terphenyl derivatives | | Descriptor: | (3~{R})-1-[[4-[2-chloranyl-3-(2,3-dihydro-1,4-benzodioxin-6-yl)phenyl]-2-methoxy-phenyl]methyl]-~{N}-(2-hydroxyethyl)pyrrolidine-3-carboxamide, CHLORIDE ION, Programmed cell death 1 ligand 1, ... | | Authors: | Plewka, J, Magiera-Mularz, K, Surmiak, E, Kalinowska-Tluscik, J, Holak, T.A. | | Deposit date: | 2023-05-12 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Solubilizer Tag Effect on PD-L1/Inhibitor Binding Properties for m -Terphenyl Derivatives.
Acs Med.Chem.Lett., 15, 2024
|
|
6A03
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6GFR
 
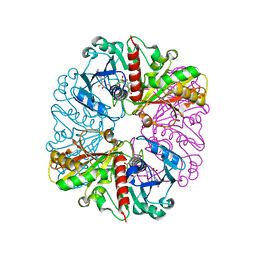 | | cyanobacterial GAPDH with NAD | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, MAGNESIUM ION, ... | | Authors: | McFarlane, C.R, Murray, J.W. | | Deposit date: | 2018-05-01 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural basis of light-induced redox regulation in the Calvin-Benson cycle in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6Y33
 
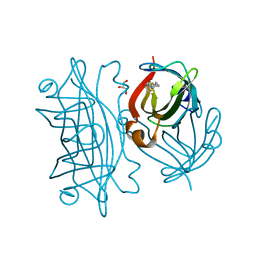 | | Streptavidin mutant S112R with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6GI3
 
 | |
6ZZE
 
 | |
8RLO
 
 | | Human Carbonic Anhydrase I in complex with veralipride | | Descriptor: | Carbonic anhydrase 1, Veralipride, (S)-, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2024-01-03 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The dopamine D 2 receptors antagonist Veralipride inhibits carbonic anhydrases: solution and crystallographic insights on human isoforms.
Chem Asian J, 19, 2024
|
|
6Y4Z
 
 | | The crystal structure of human MACROD2 in space group P43212 | | Descriptor: | L(+)-TARTARIC ACID, Thioredoxin 1,ADP-ribose glycohydrolase MACROD2 | | Authors: | Wazir, S, Maksimainen, M.M, Lehtio, L. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple crystal forms of human MacroD2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
8TR3
 
 | | Cryo-EM structure of HmAb64 scFv in complex with CNE40 SOSIP trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CNE40 SOSIP Envelope glycoprotein gp120, ... | | Authors: | Chan, K.-W, Kong, X.P. | | Deposit date: | 2023-08-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Human CD4-binding site antibody elicited by polyvalent DNA prime-protein boost vaccine neutralizes cross-clade tier-2-HIV strains.
Nat Commun, 15, 2024
|
|
8G6C
 
 | | GTP Cyclohydrolase-IB with manganese | | Descriptor: | 1,2-ETHANEDIOL, GTP cyclohydrolase FolE2, MANGANESE (II) ION | | Authors: | McWhorter, K.L, Amaya Lopez, C.Y, Davis, K.M. | | Deposit date: | 2023-02-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Combatting melioidosis with chemical synthetic lethality.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8RD4
 
 | | Telomeric RAP1:DNA-PK complex | | Descriptor: | DNA (41-MER), DNA-dependent protein kinase catalytic subunit, Telomeric repeat-binding factor 2-interacting protein 1, ... | | Authors: | Eickhoff, P, Fisher, C.E.L, Inian, O, Guettler, S, Douglas, M.E. | | Deposit date: | 2023-12-07 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Chromosome end protection by RAP1-mediated inhibition of DNA-PK.
Nature, 642, 2025
|
|
6GI1
 
 | | Crystal structure of the ferric enterobactin esterase (pfeE) mutant(S157A) from Pseudomonas aeruginosa in presence of enterobactin | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, FE (III) ION, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-05-09 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A Key Role for the Periplasmic PfeE Esterase in Iron Acquisition via the Siderophore Enterobactin in Pseudomonas aeruginosa.
ACS Chem. Biol., 13, 2018
|
|
8B54
 
 | | CDK2/cyclin A2 in complex with pyrazolo[4,3-d]pyrimidine inhibitor LGR6768 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Djukic, S, Skerlova, J, Rezacova, P. | | Deposit date: | 2022-09-21 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of new highly selective pyrazolo[4,3-d]pyrimidine inhibitor of CDK7.
Biomed Pharmacother, 161, 2023
|
|
7AOA
 
 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
8DD5
 
 | | Crystal structure of KAT6A in complex with inhibitor CTx-648 (PF-9363) | | Descriptor: | 2,6-dimethoxy-N-{4-methoxy-6-[(1H-pyrazol-1-yl)methyl]-1,2-benzoxazol-3-yl}benzene-1-sulfonamide, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Greasley, S.E, Johnson, E, Brodsky, O. | | Deposit date: | 2022-06-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Targeting KAT6A/KAT6B dependencies in breast cancer with a novel selective, orally bioavailable KAT6 inhibitor, CTx-648/PF-9363
To Be Published
|
|
6YCR
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | FFIVIRDRVFR(CCS)G(NH2), Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Grudnik, P, Kuska, K, Holak, T.A, Dubin, G. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Macrocyclic Peptide Inhibitor of PD-1/PD-L1 Immune Checkpoint
Adv. Ther., 2020
|
|
8AD9
 
 | | Crystal structure of ClpC2 C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cyclomarin A, ... | | Authors: | Taylor, G, Cui, H.J, Leodolter, J, Giese, C, Weber-Ban, E. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | ClpC2 protects mycobacteria against a natural antibiotic targeting ClpC1-dependent protein degradation.
Commun Biol, 6, 2023
|
|
