6P5P
 
 | | Discovery of a Novel, Highly Potent, and Selective Thieno[3,2-d]pyrimidinone-Based Cdc7 inhibitor with a Quinuclidine Moiety (TAK-931) as an Orally Active Investigational Anti-Tumor Agent | | Descriptor: | 2-[(2S)-1-azabicyclo[2.2.2]octan-2-yl]-6-(5-methyl-1H-pyrazol-4-yl)thieno[3,2-d]pyrimidin-4(3H)-one, Rho-associated protein kinase 2 | | Authors: | Hoffman, I.D, Skene, R.J. | | Deposit date: | 2019-05-30 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery of a Novel, Highly Potent, and Selective Thieno[3,2-d]pyrimidinone-Based Cdc7 Inhibitor with a Quinuclidine Moiety (TAK-931) as an Orally Active Investigational Antitumor Agent.
J.Med.Chem., 63, 2020
|
|
8DEK
 
 | |
7A9Y
 
 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | Cellular retinoic acid-binding protein 1, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6D5K
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase bound to ATP, and Adenosylcobalamin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-DEOXYADENOSINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dodge, G.J, Campanello, G, Smith, J.L, Banerjee, R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Sacrificial Cobalt-Carbon Bond Homolysis in Coenzyme B12as a Cofactor Conservation Strategy.
J. Am. Chem. Soc., 140, 2018
|
|
7A5Z
 
 | | Structure of VIM-2 metallo-beta-lactamase with hydrolysed Faropenem imine product | | Descriptor: | (5~{Z})-2-[1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-(4-oxidanylbutylidene)-2~{H}-1,3-thiazole-4-carboxylic acid, Beta-lactamase VIM-2, CHLORIDE ION, ... | | Authors: | Lucic, A, Schofield, C.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
7A60
 
 | | Crystal structure of VIM-2 with hydrolyzed faropenem (ring-open form) | | Descriptor: | (5~{Z})-2-[1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-5-(4-oxidanylbutylidene)-2~{H}-1,3-thiazole-4-carboxylic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Faropenem reacts with serine and metallo-beta-lactamases to give multiple products.
Eur.J.Med.Chem., 215, 2021
|
|
5INL
 
 | | Mouse Tdp2 reaction product (5'-phosphorylated DNA)-Mg2+ complex with deoxyadenosine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, DNA (5'-D(P*AP*CP*GP*AP*AP*TP*TP*CP*G)-3'), ... | | Authors: | Schellenberg, M.J, Vilas, C.K, Williams, R.S. | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2.
Nucleic Acids Res., 44, 2016
|
|
7ZPR
 
 | | KtrAB complex with N-terminal deletion of KtrB 1-19 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Vonck, J, Stautz, J. | | Deposit date: | 2022-04-28 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | KtrAB complex with N-terminal deletion of KtrB 1-19
To Be Published
|
|
7ZI1
 
 | | Crystal structure of dCK C4S-S74E mutant in complex with UDP and the dCKi1 inhibitor | | Descriptor: | Deoxycytidine kinase, N-[3-[[4-(4-azanylpyrimidin-2-yl)-1,3-thiazol-2-yl]amino]-4-methyl-phenyl]-4-[(4-methylpiperazin-1-yl)methyl]benzamide, SODIUM ION, ... | | Authors: | Saez-Ayala, M, Ben-Yaala, K, Betzi, S, Rebuffet, E, Morelli, X. | | Deposit date: | 2022-04-07 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia.
Nat Commun, 14, 2023
|
|
8ZFS
 
 | |
8WFR
 
 | | The Crystal Structure of PCSK9 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Lu, Y. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of PCSK9 from Biortus.
To Be Published
|
|
7RPY
 
 | | X25-2 domain of Sca5 from Ruminococcus bromii | | Descriptor: | ACETATE ION, Cohesin-containing protein, GLYCEROL, ... | | Authors: | Cerqueira, F, Koropatkin, N. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Sas20 is a highly flexible starch-binding protein in the Ruminococcus bromii cell-surface amylosome.
J.Biol.Chem., 298, 2022
|
|
6XWB
 
 | |
7ZIA
 
 | | Crystal structure of dCK C4S-S74E mutant in complex with UDP and the OR0634 inhibitor | | Descriptor: | 2-[2-[[2-methyl-5-[6-(4-methylpiperazin-1-yl)sulfonylpyridin-3-yl]phenyl]-propyl-amino]-1,3-thiazol-4-yl]pyrimidine-4,6-diamine, Deoxycytidine kinase, SODIUM ION, ... | | Authors: | Ben-Yaala, K, Saez-Ayala, M, Betzi, S, Rebuffet, E, Morelli, X. | | Deposit date: | 2022-04-07 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia.
Nat Commun, 14, 2023
|
|
7ZI2
 
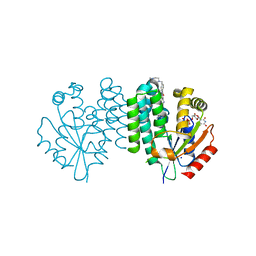 | | Crystal structure of dCK C4S-S74E mutant in complex with UDP and the dCKi2 inhibitor | | Descriptor: | Deoxycytidine kinase, N-[3-[[4-[4,6-bis(azanyl)pyrimidin-2-yl]-1,3-thiazol-2-yl]amino]-4-methyl-phenyl]-4-[(4-methylpiperazin-1-yl)methyl]benzamide, SODIUM ION, ... | | Authors: | Saez-Ayala, M, Ben-Yaala, K, Betzi, S, Rebuffet, E, Morelli, X. | | Deposit date: | 2022-04-07 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | From a drug repositioning to a structure-based drug design approach to tackle acute lymphoblastic leukemia.
Nat Commun, 14, 2023
|
|
8HY0
 
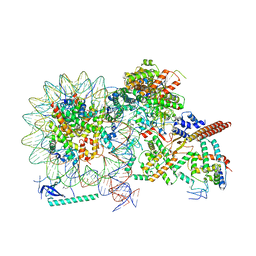 | |
8TKD
 
 | | Human Type 3 IP3 Receptor - Preactivated State (+IP3/ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8HXX
 
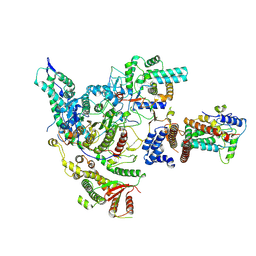 | | Cryo-EM structure of the histone deacetylase complex Rpd3S | | Descriptor: | Chromatin modification-related protein EAF3, Histone H3, Histone deacetylase RPD3, ... | | Authors: | Cui, H, Wang, H. | | Deposit date: | 2023-01-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of histone deacetylase complex Rpd3S bound to nucleosome.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7A8S
 
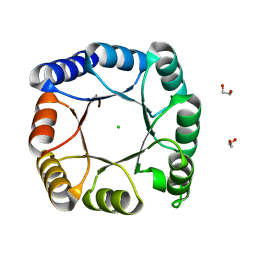 | | de novo designed ba8-barrel sTIM11 with an alpha-helical extension | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, sTIM11_h3 | | Authors: | Shanmugaratnam, S, Wiese, J.G, Hocker, B. | | Deposit date: | 2020-08-31 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Extension of a de novo TIM barrel with a rationally designed secondary structure element.
Protein Sci., 30, 2021
|
|
9J9K
 
 | |
8TKI
 
 | | Human Type 3 IP3 Receptor - Labile Resting State 2 (+IP3/ATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8A1N
 
 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with fumaryl amide analogue 13 | | Descriptor: | (Z)-N-(4-chlorophenyl)-4-oxidanylidene-but-2-enamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | de Munnik, M, Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-throughput screen with the l,d-transpeptidase Ldt Mt2 of Mycobacterium tuberculosis reveals novel classes of covalently reacting inhibitors.
Chem Sci, 14, 2023
|
|
8TKE
 
 | | Human Type 3 IP3 Receptor - Preactivated+Ca2+ State (+IP3/ATP/JD Ca2+) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Paknejad, N, Sapuru, V, Hite, R.K. | | Deposit date: | 2023-07-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural titration reveals Ca 2+ -dependent conformational landscape of the IP 3 receptor.
Nat Commun, 14, 2023
|
|
8D5N
 
 | | Crystal structure of Ld-HF10 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2022-06-05 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide Centric V beta Specific Germline Contacts Shape a Specialist T Cell Response.
Front Immunol, 13, 2022
|
|
7PE9
 
 | |
