8YBC
 
 | |
6RVC
 
 | | Crystal structure of Patched-1 ectodomain 2 (PTCH1-ECD2) in complex with nanobody 75 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody NB75, Protein patched homolog 1, ... | | Authors: | Rudolf, A.F, Kowatsch, C, El Omari, K, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
1EVB
 
 | |
6ENI
 
 | | Tryptophan Repressor TrpR from E.coli variant T44L S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, Trp operon repressor | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-10-04 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
6ZC2
 
 | | Crystal structure of RahU protein in complex with TRIS molecule | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RahU protein | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of RahU, an aegerolysin protein from the human pathogen Pseudomonas aeruginosa, and its interaction with membrane ceramide phosphorylethanolamine.
Sci Rep, 11, 2021
|
|
8T2H
 
 | | DYRK1A complex with DYR530 | | Descriptor: | (4P)-4-{(3M)-3-[3-fluoro-4-(4-methylpiperazin-1-yl)phenyl]-2-methyl-3H-imidazo[4,5-b]pyridin-5-yl}pyridin-2-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, GLYCEROL, ... | | Authors: | Montfort, W.R, Basantes, L.E. | | Deposit date: | 2023-06-06 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Functional Characterization of a Potent, Selective, and Metabolically Stable PROTAC of the Protein Kinases DYRK1A and DYRK1B.
J.Med.Chem., 67, 2024
|
|
6QEE
 
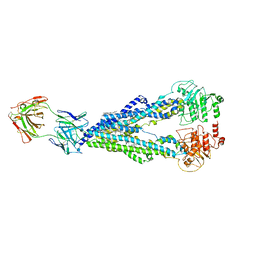 | |
9J5X
 
 | | Cryo-EM Structure of URAT1 in Complex with Lingolinurad | | Descriptor: | 3-bromanyl-5-(2-ethylimidazo[1,2-a]pyridin-3-yl)carbonyl-2-oxidanyl-benzenecarbonitrile, Solute carrier family 22 member 12 | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2024-08-13 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural Basis for Inhibition of Urate Reabsorption in URAT1.
Jacs Au, 5, 2025
|
|
9HIS
 
 | |
6E9L
 
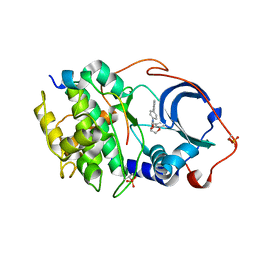 | |
5AQ8
 
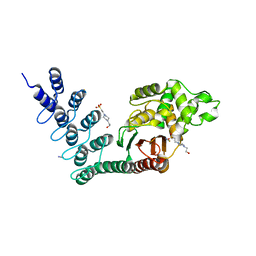 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, OFF7_DB12V4, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
9J5W
 
 | | Cryo-EM Structure of URAT1 in Complex with Benzbromarone | | Descriptor: | Solute carrier family 22 member 12, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2024-08-13 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis for Inhibition of Urate Reabsorption in URAT1.
Jacs Au, 5, 2025
|
|
1EVA
 
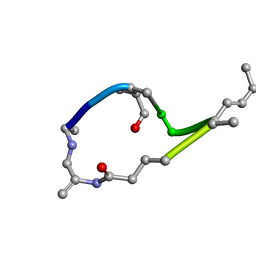 | |
1EVD
 
 | |
8F97
 
 | | Compound 5 bound to procaspase-6 | | Descriptor: | 1,2-ETHANEDIOL, 2-({[(2M)-[2,3'-bipyridin]-2'-yl]amino}methyl)-5-fluorophenol, CHLORIDE ION, ... | | Authors: | Fan, P, Zhao, Y, Renslo, A.R, Arkin, M.R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Systematic Study of Heteroarene Stacking Using a Congeneric Set of Molecular Glues for Procaspase-6.
J.Med.Chem., 66, 2023
|
|
8F39
 
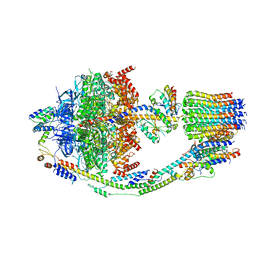 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6EI2
 
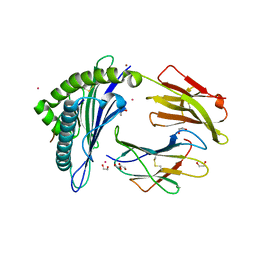 | | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Picaud, S, Guillaume, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gfeller, D, Filippakopoulos, P. | | Deposit date: | 2017-09-16 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide
To Be Published
|
|
4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
5IX8
 
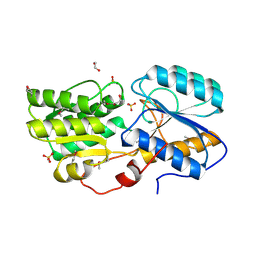 | | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822 | | Descriptor: | 1,2-ETHANEDIOL, Putative sugar ABC transport system, substrate-binding protein, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-23 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822
To Be Published
|
|
6PF1
 
 | |
5IAU
 
 | |
5IEH
 
 | |
6BRL
 
 | |
4U5X
 
 | | Structure of plant small GTPase OsRac1 complexed with the non-hydrolyzable GTP analog GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ohki, I, Kosami, K, Fujiwara, T, Nakagawa, A, Shimamoto, K, Kojima, C. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of the Plant Small GTPase OsRac1 Reveals Its Mode of Binding to NADPH Oxidase
J.Biol.Chem., 289, 2014
|
|
6I9Q
 
 | | Structure of the mouse CD98 heavy chain ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, CHLORIDE ION | | Authors: | Schiefner, A, Deuschle, F.-C, Skerra, A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural differences between the ectodomains of murine and human CD98hc.
Proteins, 87, 2019
|
|
