7T8R
 
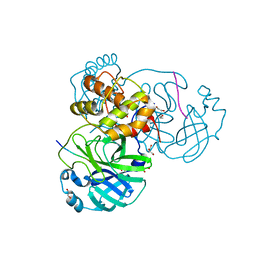 | |
6N6S
 
 | | Crystal structure of ABIN-1 UBAN | | Descriptor: | TNFAIP3-interacting protein 1 | | Authors: | Rahighi, S, Dikic, I, Wakatsuki, S. | | Deposit date: | 2018-11-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Recognition of M1-Linked Ubiquitin Chains by Native and Phosphorylated UBAN Domains.
J.Mol.Biol., 431, 2019
|
|
6N92
 
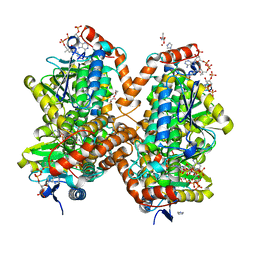 | | Methylmalonyl-CoA decarboxylase in complex with 2-nitronate-propionyl-CoA | | Descriptor: | (2E)-2-(hydroxyimino)propanoic acid, IMIDAZOLE, Methylmalonyl-CoA decarboxylase, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
7OTV
 
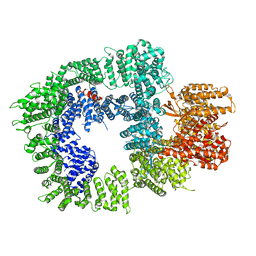 | | DNA-PKcs in complex with wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
9B1I
 
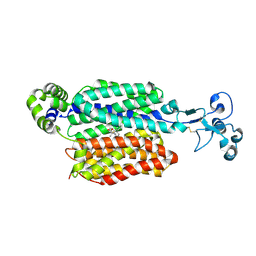 | | Human urate transporter 1 URAT1 in complex with verinurad | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 22 member 12, verinurad | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 34, 2024
|
|
8UL1
 
 | |
9KFU
 
 | | Human FGFR3 in complex with inhibitor F1 | | Descriptor: | (7~{S})-2-azanyl-7-(4-fluorophenyl)-6,7-dihydro-4~{H}-[1,3]thiazolo[4,5-b]pyridin-5-one, COBALT (II) ION, Fibroblast growth factor receptor 3, ... | | Authors: | Zhang, J, Song, K, Li, M.Y. | | Deposit date: | 2024-11-07 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Electron-density-informed effective and reliable de novo molecular design and optimization with ED2Mol
Nat Mach Intell, 7, 2025
|
|
6ZRN
 
 | | Crystal structure of the RLIP76 Ral binding domain mutant (E427S/L429M/Q433L/K440R) in complex with RalB-GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hurd, C, Brear, P, Revell, J, Ross, S, Mott, H, Owen, D. | | Deposit date: | 2020-07-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Affinity maturation of the RLIP76 Ral binding domain to inform the design of stapled peptides targeting the Ral GTPases.
J.Biol.Chem., 296, 2020
|
|
6X7N
 
 | | LnmK in complex with 2-nitronate-propionyl-amino(dethia)-CoA | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP acyltransferase/decarboxylase, SULFATE ION, [1-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylamino]-1-oxidanylidene-propan-2-ylidene]-bis(oxidanidyl)azanium | | Authors: | Stunkard, L.M, Kick, B.J, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structures of LnmK, a Bifunctional Acyltransferase/Decarboxylase, with Substrate Analogues Reveal the Basis for Selectivity and Stereospecificity.
Biochemistry, 60, 2021
|
|
8QHQ
 
 | | Crystal structure of human DNPH1 bound to hmdUMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
7A6E
 
 | |
9IIO
 
 | |
8XW3
 
 | |
8SBM
 
 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | Descriptor: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | Authors: | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | Deposit date: | 2023-04-03 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
8XPX
 
 | | The Crystal Structure of PARP12 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Shen, Z. | | Deposit date: | 2024-01-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of PARP12 from Biortus.
To Be Published
|
|
8AWZ
 
 | |
6XJ8
 
 | | KPC-2 N170A mutant bound to hydrolyzed imipenem at 2.05 A | | Descriptor: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2020-06-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | KPC-2 beta-lactamase enables carbapenem antibiotic resistance through fast deacylation of the covalent intermediate.
J.Biol.Chem., 296, 2020
|
|
8XS4
 
 | |
8XW2
 
 | |
6OUK
 
 | |
8XS5
 
 | |
8EYJ
 
 | | Crystal Structure of uncleaved SARS-CoV-2 Main Protease C145S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | An in-solution snapshot of SARS-COV-2 main protease maturation process and inhibition.
Nat Commun, 14, 2023
|
|
5M3T
 
 | | Structural tuning of CD81LEL (space group P64) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, CHLORIDE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.021 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
8XVX
 
 | |
8PJ8
 
 | | FKBP51FK1 F67E/K60Orn (i, i+7) in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Charalampidou, A, Hausch, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
