8VDA
 
 | |
8BZ5
 
 | |
7BQ2
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, 15-meric peptide from Nuclear receptor coactivator 1, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-03-23 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
8VD9
 
 | |
8V8C
 
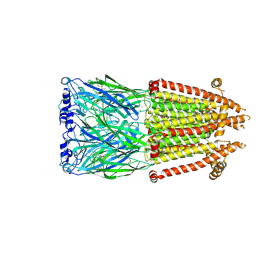 | | Alpha7-nicotinic acetylcholine receptor time resolved bound to epibatidine and PNU-120596 asymmetric state 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
6X3C
 
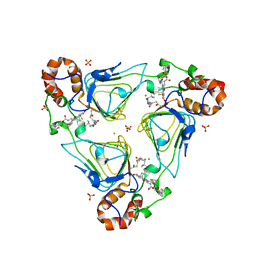 | | Crystal structure of streptogramin A acetyltransferase VatA from Staphylococcus aureus in complex with streptogramin analog F1037 (47) | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaires, H.A, Fraser, J.S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
1A54
 
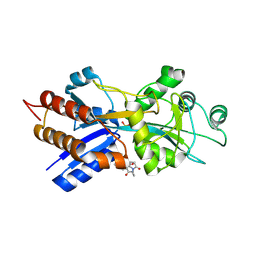 | | PHOSPHATE-BINDING PROTEIN MUTANT A197C LABELLED WITH A COUMARIN FLUOROPHORE AND BOUND TO DIHYDROGENPHOSPHATE ION | | Descriptor: | DIHYDROGENPHOSPHATE ION, N-[2-(1-MALEIMIDYL)ETHYL]-7-DIETHYLAMINOCOUMARIN-3-CARBOXAMIDE, Phosphate-binding protein PstS | | Authors: | Hirshberg, M, Henrick, K, Lloyd-Haire, L, Vasisht, N, Brune, M, Corrie, J.E.T, Webb, M.R. | | Deposit date: | 1998-02-19 | | Release date: | 1998-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate.
Biochemistry, 37, 1998
|
|
7RFR
 
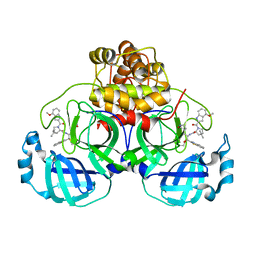 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-(4-methoxy-1H-indole-2-carbonyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
8H2X
 
 | | Structure of Acb2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
7BU0
 
 | |
7RQ8
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
8X4A
 
 | |
8BVX
 
 | |
7BYL
 
 | | Cryo-EM structure of human KCNQ4 | | Descriptor: | Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, POTASSIUM ION, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
6QP5
 
 | |
4RVO
 
 | |
8X4C
 
 | |
6OAH
 
 | |
8HUA
 
 | | Serial synchrotron crystallography structure of ba3-type cytochrome c oxidase from Thermus thermophilus using a goniometer compatible flow-cell | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Ghosh, S, Zoric, D, Bjelcic, M, Johannesson, J, Sandelin, E, Branden, G, Neutze, R. | | Deposit date: | 2022-12-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A simple goniometer-compatible flow cell for serial synchrotron X-ray crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
8BM8
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 11 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(3-cyanophenyl)-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-10 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | (placeholder, will be filled after publication)
To Be Published
|
|
8X4B
 
 | |
6O7L
 
 | |
6Z8C
 
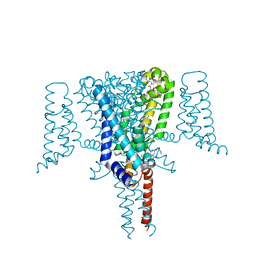 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with N-desmethyltamoxifen (3.2 A resolution) | | Descriptor: | 2-[4-[(~{Z})-1,2-diphenylbut-1-enyl]phenoxy]-~{N}-methyl-ethanamine, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2020-06-02 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
8X4E
 
 | |
8X4D
 
 | |
