9EWC
 
 | | DNA Polymerase Lambda I493R 528-530 NEY, TTP:At Ca2+ Ground State Ternary Complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA polymerase lambda, ... | | Authors: | Nourisson, A, Haouz, A, Missoury, S, Delarue, M. | | Deposit date: | 2024-04-03 | | Release date: | 2025-07-23 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (3.67 Å) | | Cite: | Fidelity, specialization, and evolution of Paramecium PolX DNA polymerases involved in programmed double-strand break DNA repair.
Nucleic Acids Res., 53, 2025
|
|
4PZN
 
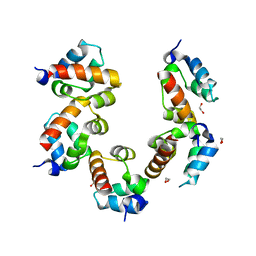 | | Crystal structure of PHC3 SAM L971E | | Descriptor: | 1,2-ETHANEDIOL, Polyhomeotic-like protein 3 | | Authors: | Nanyes, D.R, Junco, S.E, Taylor, A.B, Robinson, A.K, Patterson, N.L, Shivarajpur, A, Halloran, J, Hale, S.M, Kaur, Y, Hart, P.J, Kim, C.A. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple polymer architectures of human polyhomeotic homolog 3 sterile alpha motif.
Proteins, 82, 2014
|
|
7A55
 
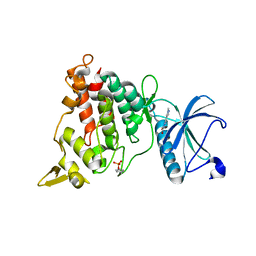 | | Structure of DYRK1A in complex with compound 8 | | Descriptor: | 3-(1-methylpyrazol-4-yl)-1~{H}-pyrazole-5-carboxylic acid, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
6VCV
 
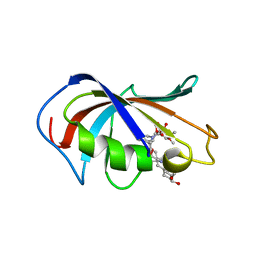 | | Aspergillus fumigatus FKBP12 protein bound with APX879 in P1 space group | | Descriptor: | FK506-binding protein 1A, N'-[(3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,26aS)-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-1,20,21-trioxo-8-(prop-2-en-1-yl)-1,3,4,5,6,8,11,12,13,14,15,16,17,18,19,20,21,23,24,25,26,26a-docosahydro-7H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosin-7-ylidene]acetohydrazide | | Authors: | Gobeil, S, Spicer, L. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Leveraging Fungal and Human Calcineurin-Inhibitor Structures, Biophysical Data, and Dynamics To Design Selective and Nonimmunosuppressive FK506 Analogs.
Mbio, 12, 2021
|
|
6VD1
 
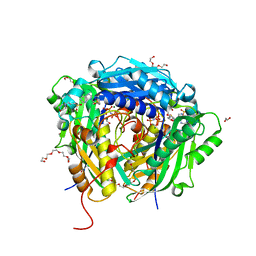 | | Crystal structure of Arabidopsis thaliana S-adenosylmethionine Synthase 2 (AtMAT2) in complex with S-adenosylmethionine and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
9EWG
 
 | | DNA Polymerase Lambda I493K, TTP:At Ca2+ Ground State Ternary Complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA polymerase lambda, ... | | Authors: | Nourisson, A, Haouz, A, Missoury, S, Delarue, M. | | Deposit date: | 2024-04-03 | | Release date: | 2025-07-23 | | Last modified: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fidelity, specialization, and evolution of Paramecium PolX DNA polymerases involved in programmed double-strand break DNA repair.
Nucleic Acids Res., 53, 2025
|
|
4Q0J
 
 | | Deinococcus radiodurans BphP photosensory module | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | Crystallographic and Electron Microscopic Analyses of a Bacterial Phytochrome Reveal Local and Global Rearrangements during Photoconversion.
J.Biol.Chem., 289, 2014
|
|
6ZQK
 
 | | HER2-binding scFv-Fab fusion 841 | | Descriptor: | 1,2-ETHANEDIOL, 841 heavy chain, 841 light chain | | Authors: | Kast, F, Schwill, M, Stueber, J.C, Pfundstein, S, Nagy-Davidescu, G, Monne Rodriguez, J.M, Seehusen, F, Richter, C.P, Honegger, A, Hartmann, K.P, Weber, T.G, Kroener, F, Ernst, P, Piehler, J, Plueckthun, A. | | Deposit date: | 2020-07-09 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineering an anti-HER2 biparatopic antibody with a multimodal mechanism of action.
Nat Commun, 12, 2021
|
|
6MVQ
 
 | | HCV NS5B 1b N316 bound to Compound 31 | | Descriptor: | (4-{1-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-1H-1,2,4-triazol-5-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
6XIY
 
 | | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to methyl 2-(acetylamino)-2-deoxy-1-thio-alpha-D-galactopyranose | | Descriptor: | C-type lectin domain family 10 member A, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Birrane, G, Murphy, P.V, Gabba, A, Luz, J.G. | | Deposit date: | 2020-06-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to GalNAc and the Tumor-Associated Tn Antigen.
Biochemistry, 60, 2021
|
|
5IH8
 
 | | MELK in complex with NVS-MELK1 | | Descriptor: | Maternal embryonic leucine zipper kinase, N-[(pyridin-2-yl)methyl]-4-[4-(pyridin-4-yl)-1H-pyrazol-1-yl]benzamide | | Authors: | Sprague, E.R, Puleo, D.E. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
5LHP
 
 | | The p-aminobenzamidine active site inhibited catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment, P-AMINO BENZAMIDINE, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P.J, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
8WCQ
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
5LPB
 
 | |
7OSE
 
 | | cytochrome bd-II type oxidase with bound aurachin D | | Descriptor: | Aurachin D, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-II ubiquinol oxidase subunit 1, ... | | Authors: | Grauel, A, Kaegi, J, Rasmussen, T, Wohlwend, D, Boettcher, B, Friedrich, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Escherichia coli cytochrome bd-II type oxidase with bound aurachin D.
Nat Commun, 12, 2021
|
|
8U5M
 
 | | Structure of Sts-1 HP domain with rebamipide | | Descriptor: | Rebamipide, Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Azia, F, Dey, R, French, J.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Rebamipide and Derivatives are Potent, Selective Inhibitors of Histidine Phosphatase Activity of the Suppressor of T Cell Receptor Signaling Proteins.
J.Med.Chem., 67, 2024
|
|
5IOH
 
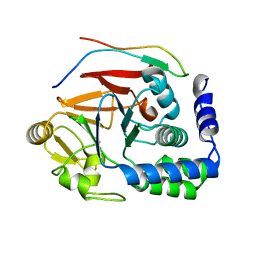 | | RepoMan-PP1a (protein phosphatase 1, alpha isoform) holoenzyme complex | | Descriptor: | Cell division cycle-associated protein 2, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-08 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
8AHE
 
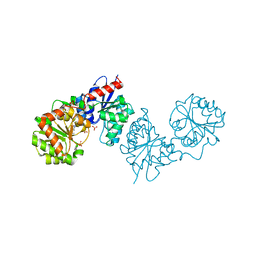 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
6ZMV
 
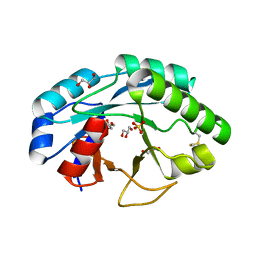 | | Structure of muramidase from Trichobolus zukalii | | Descriptor: | GLYCEROL, SULFATE ION, muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
8AHF
 
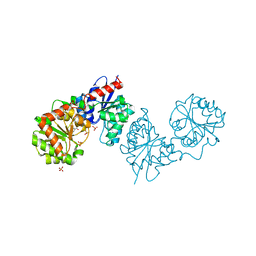 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
4Q6B
 
 | | Crystal Structure of ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense complex with Leu | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Extracellular ligand-binding receptor, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | Crystal Structure of ABC Transporter Substrate-Binding Protein fromDesulfitobacterium hafniense complex with Leu
To be Published, 2014
|
|
9EYG
 
 | | Structure of Tannerella forsythia endopeptidase O (TfPepO) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zak, K.M, Grudnik, P, Waligorska, I, Ksiazek, M. | | Deposit date: | 2024-04-09 | | Release date: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Tannerella forsythia endopeptidase O (TfPepO)
To Be Published
|
|
7R5C
 
 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-29 (G206C, R207S, D210L, S211V) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Isoaspartyl peptidase, ... | | Authors: | Barciszewski, J, Imiolczyk, B, Loch, J.I, Jaskolski, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8WCR
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
6V9E
 
 | | The crystal structure of the 2009 H1N1 PA endonuclease wild type in complex with SJ000988632 | | Descriptor: | 1,1',1'',1''',1''''-[(3R,5S,7S,9R)-decane-1,3,5,7,9-pentayl]penta(pyrrolidin-2-one), MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-13 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
