8E8L
 
 | | 9H2 Fab-poliovirus 1 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
6QU8
 
 | | Adenovirus Serotype 26 (Ad26) in complex with sialic acid, pH8.0 | | Descriptor: | 1,2-ETHANEDIOL, Fiber, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Baker, A.T, Rizkallah, P.J, Parker, A.L, Mundy, R.M. | | Deposit date: | 2019-02-26 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Human adenovirus type 26 uses sialic acid-bearing glycans as a primary cell entry receptor.
Sci Adv, 5, 2019
|
|
7PVU
 
 | | Crystal structure of p38alpha C162S in complex with CAS2094511-69-8, P 1 21 1 | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-cyclobutyl-1H-1,3-benzodiazol-5-yl)-2-fluorobenzene-1-sulfonamide | | Authors: | Baginski, B, Pous, J, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2021-10-05 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Characterization of p38 alpha autophosphorylation inhibitors that target the non-canonical activation pathway.
Nat Commun, 14, 2023
|
|
4RB4
 
 | | Crystal structure of dodecameric iron-containing heptosyltransferase TibC in complex with ADP-D-beta-D-heptose at 3.9 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Glycosyltransferase tibC, ... | | Authors: | Yao, Q, Lu, Q, Shao, F. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | A structural mechanism for bacterial autotransporter glycosylation by a dodecameric heptosyltransferase family
Elife, 3, 2014
|
|
7S1C
 
 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001897 (compound 1) | | Descriptor: | COPPER (II) ION, Thiol:disulfide interchange protein DsbA, ~{N}-methyl-1-(3-thiophen-3-ylphenyl)methanamine | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Sharma, P. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
7S1F
 
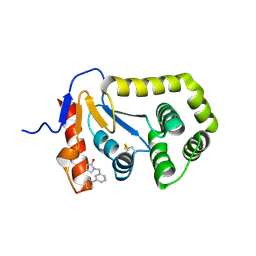 | | Crystal structure of E.coli DsbA in complex with compound MIPS-0001886 (compound 38) | | Descriptor: | 1-[(3-thiophen-3-ylphenyl)methyl]-3~{H}-pyrrol-2-one, COPPER (II) ION, GLYCEROL, ... | | Authors: | Heras, B, Scanlon, M.J, Martin, J.L, Caria, S. | | Deposit date: | 2021-09-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fluoromethylketone-fragment conjugates designed as covalent modifiers of EcDsbA are atypical substrates
Chemrxiv, 2022
|
|
8JES
 
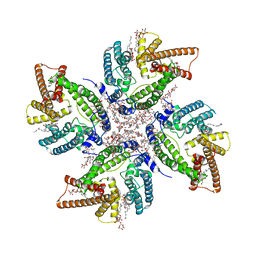 | | Cryo-EM structure of DltB homo-tetramer | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, DIACYL GLYCEROL, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Zhang, P, Liu, Z. | | Deposit date: | 2023-05-16 | | Release date: | 2024-05-22 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural insights into the transporting and catalyzing mechanism of DltB in LTA D-alanylation.
Nat Commun, 15, 2024
|
|
8UQU
 
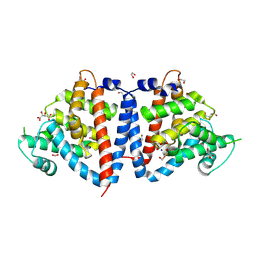 | | Crystal Structure of N-terminal Domain of Fic Family Protein from Bordetella bronchiseptica | | Descriptor: | 1,2-ETHANEDIOL, D(-)-TARTARIC ACID, Fido domain-containing protein, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-24 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of N-terminal Domain of Fic Family Protein from Bordetella bronchiseptica
To Be Published
|
|
6YG1
 
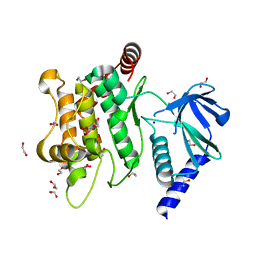 | | Crystal structure of MKK7 (MAP2K7) in an active state, allosterically triggered by the N-terminal helix | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, SODIUM ION | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC), Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
8R4U
 
 | | Structure of salt-inducible kinase 3 with inhibitors | | Descriptor: | 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-fluoranylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, SULFATE ION, Serine/threonine-protein kinase SIK3, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2023-11-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | The structures of salt-inducible kinase 3 in complex with inhibitors reveal determinants for binding and selectivity.
J.Biol.Chem., 300, 2024
|
|
9GDV
 
 | | Highly optimized CNS penetrant inhibitors of EGFR Exon20 Insertion Mutations | | Descriptor: | Epidermal growth factor receptor, SULFATE ION, ~{N}-[2-[2-(dimethylamino)ethyl-methyl-amino]-4-methoxy-5-[[4-(1-methylindol-3-yl)pyrimidin-2-yl]amino]phenyl]propanamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-08-06 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Highly Optimized CNS Penetrant Inhibitors of EGFR Exon20 Insertion Mutations.
J.Med.Chem., 68, 2025
|
|
9B8E
 
 | | Structure of S-nitrosylated Legionella pneumophila Ceg10. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tomchick, D.R, Heisler, D.B, Alto, N.M. | | Deposit date: | 2024-03-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploiting bacterial effector proteins to uncover evolutionarily conserved antiviral host machinery.
Plos Pathog., 20, 2024
|
|
9E1K
 
 | | Discovery of Potent, Highly Selective and Efficacious SMARCA2 Degraders - Compound 11 | | Descriptor: | (12S)-4-bromo-7,7-dimethyl-9-(piperidin-4-yl)indolo[1,2-a]quinazolin-5(7H)-one, Isoform Short of Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2024-10-21 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
J.Med.Chem., 68, 2025
|
|
1AGU
 
 | | THE SOLUTION NMR STRUCTURE OF THE C10R ADDUCT OF BENZO[A]PYRENE-DIOL-EPOXIDE AT THE N6 POSITION OF ADENINE OF AN 11 BASE-PAIR OLIGONUCLEOTIDE SEQUENCE CODING FOR AMINO ACIDS 60-62 OF THE PRODUCT OF THE N-RAS PROTOONCOGENE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*GP*GP*AP*CP*EP*AP*GP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Zegar, I.S, Stone, M.P. | | Deposit date: | 1997-03-25 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Adduction of the human N-ras codon 61 sequence with (-)-(7S,8R,9R,10S)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a] pyrene: structural refinement of the intercalated SRSR(61,2) (-)-(7S,8R,9S,1 0R)-N6-[10-(7,8,9,10- tetrahydrobenzo[a]pyrenyl)]-2'-deoxyadenosyl adduct from 1H NMR
Biochemistry, 35, 1996
|
|
8JEM
 
 | | DltB tetramer in complex with inhibitor m-AMSA | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, DIACYL GLYCEROL, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Zhang, P, Liu, Z. | | Deposit date: | 2023-05-16 | | Release date: | 2024-05-15 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into the transporting and catalyzing mechanism of DltB in LTA D-alanylation.
Nat Commun, 15, 2024
|
|
3R3K
 
 | | Crystal structure of a parallel 6-helix coiled coil | | Descriptor: | 1,2-ETHANEDIOL, CChex-Phi22 helix, CHLORIDE ION, ... | | Authors: | Zaccai, N.R, Chi, B.H.C, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.2009 Å) | | Cite: | A de novo peptide hexamer with a mutable channel.
Nat.Chem.Biol., 7, 2011
|
|
7OLU
 
 | |
8UAG
 
 | |
6HPO
 
 | |
8AHQ
 
 | | VirD/holo-ACP5b of Streptomyces virginiae complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-PHOSPHOPANTETHEINE, CHLORIDE ION, ... | | Authors: | Collin, S, Gruez, A. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
4ZIN
 
 | | Genetically encoded Phenyl Azide Photochemistry Drive Positive and Negative Functional Modulation of a Red Fluorescent Protein | | Descriptor: | 1,2-ETHANEDIOL, MCherry fluorescent protein, SULFATE ION | | Authors: | Reddington, S.C, Driezis, S, Hartley, A.M, Watson, P.D, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-04-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Genetically encoded phenyl azide photochemistry drives positive and negative functional modulation of a red fluorescent protein
Rsc Adv, 5, 2015
|
|
9BZB
 
 | | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, SULFATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-05-24 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of metallo-hydrolase-like_MBL-fold protein from Salmonella typhimurium LT2
To Be Published
|
|
8OFX
 
 | | Molecular Mechanism of trypanosomal AQP2 | | Descriptor: | Aquaglyceroporin 2, [(2~{R},4~{S})-2-[4-[[4,6-bis(azanyl)-1,3,5-triazin-2-yl]amino]phenyl]-1,3,2-dithiarsolan-4-yl]methanol | | Authors: | Weyand, S.N, Matusevicius, M, Yamashita, K. | | Deposit date: | 2023-03-17 | | Release date: | 2024-10-02 | | Last modified: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights from aquaporin structures into drug-resistant sleeping sickness
Elife, 2025
|
|
8JK0
 
 | | Crystal structure of QL-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8JJU
 
 | | Crystal structure of QD-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
