7Z0Q
 
 | | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange | | Descriptor: | 1,2-ETHANEDIOL, CLIP peptide, HLA class II histocompatibility antigen, ... | | Authors: | Roske, Y, Abualrous, E.T, Freund, C. | | Deposit date: | 2022-02-23 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MHC-II dynamics are maintained in HLA-DR allotypes to ensure catalyzed peptide exchange.
Nat.Chem.Biol., 19, 2023
|
|
6NVR
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in Apo form | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V.M, Blundell, T.L. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.562 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
8S6K
 
 | |
7Z0I
 
 | | human PEX13 SH3 domain | | Descriptor: | 1,2-ETHANEDIOL, Peroxisomal membrane protein PEX13, ZINC ION | | Authors: | Gaussmann, S, Zak, K, Sattler, M. | | Deposit date: | 2022-02-23 | | Release date: | 2023-03-08 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of peroxisomal import by the PEX13 SH3 domain and a proximal FxxxF binding motif.
Nat Commun, 15, 2024
|
|
6Y4A
 
 | | Crystal structure of the M295I variant of Ssl1 | | Descriptor: | COPPER (II) ION, Copper oxidase | | Authors: | Mielenbrink, S, Olbrich, A, Urlacher, V, Span, I. | | Deposit date: | 2020-02-20 | | Release date: | 2021-09-01 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Substitution of the axial Type 1 Cu Ligand Afford Binding of a Water Molecule in Axial Position Affecting Kinetics, Spectral, and Structural Properties of the Small Laccase Ssl1.
Chemistry, 2024
|
|
6VTS
 
 | |
6VTQ
 
 | |
8BAT
 
 | | Geobacter lovleyi NADAR | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Geobacter lovleyi NADAR | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
8ZTY
 
 | |
8BAS
 
 | | E. coli C7 DarT1 in complex with carba-NAD and DNA | | Descriptor: | 1,2-ETHANEDIOL, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DNA (5'-D(*AP*AP*GP*AP*C)-3'), ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
7ANA
 
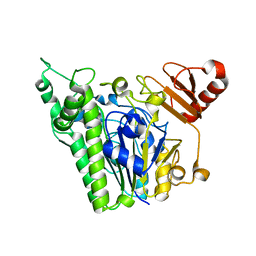 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
6NJI
 
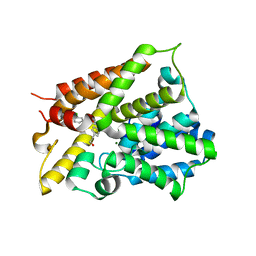 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-49 | | Descriptor: | 2-(4-{[4-(3-chlorophenyl)-6-ethyl-1,3,5-triazin-2-yl]amino}phenyl)ethan-1-ol, MAGNESIUM ION, ZINC ION, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2019-01-03 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders.
J.Med.Chem., 62, 2019
|
|
8V0C
 
 | | Structure of TDP1 catalytic domain complexed with compound IB06 | | Descriptor: | (8M)-8-(2-{[2-(fluorosulfonyl)ethyl]amino}phenyl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structures of TDP1 complexed with inhibitors
To Be Published
|
|
9AYD
 
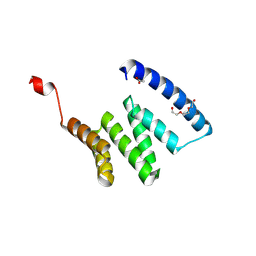 | |
8UZZ
 
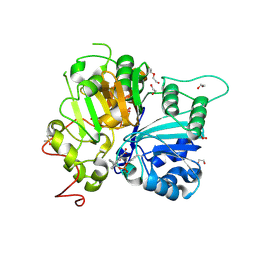 | | Structure of TDP1 catalytic domain complexed with compound IB03 | | Descriptor: | (8M)-8-{2-[(fluorosulfonyl)oxy]phenyl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Barakat, I, Wang, W, Agama, K, Al Mahmud, M.R, Pommier, Y, Burke Jr, T.R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structureal analysis of TDP1 in complex with inhbitors
To Be Published
|
|
8PMW
 
 | |
6YZ5
 
 | | H11-D4 complex with SARS-CoV-2 RBD | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Naismith, J.H, Huo, J, Mikolajek, H, Ward, P, Dumoux, M, Owens, R.J, Le Bas, A. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | H11-D4 complex with SARS-CoV-2 RBD
To Be Published
|
|
7AYW
 
 | |
7AZV
 
 | |
7SIF
 
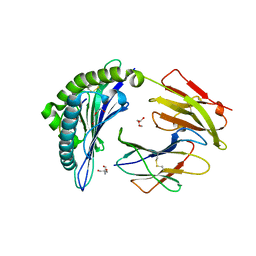 | | Crystal Structure of HLA B*3505 in complex with NPDIVIYQY, an 9-mer epitope from HIV-I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, ... | | Authors: | Gras, S, Lobos, C.A, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular insights into the HLA-B35 molecules' classification associated with HIV control.
Immunol.Cell.Biol., 102, 2024
|
|
6HQC
 
 | |
7B0E
 
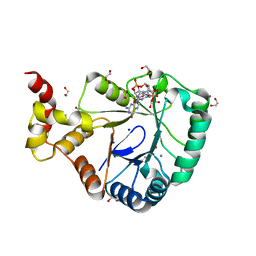 | | Crystal structure of SmbA loop deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, ... | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structure of loop deletion mutant SmbA bound to a monomeric c-di-GMP
To Be Published
|
|
8G3A
 
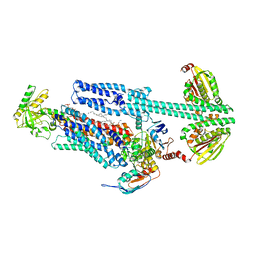 | | BceAB-S nucleotide free TM state 1 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8J9D
 
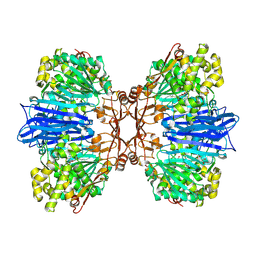 | | Crystal structure of M61 peptidase (bestatin-bound) from Xanthomonas campestris | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Yadav, P, Kumar, A, Kulkarni, B.S, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a newly identified M61 family aminopeptidase with broad substrate specificity that is solely responsible for recycling acidic amino acids.
Febs J., 291, 2024
|
|
5MCU
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LHG2) | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DNA, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
