8JSM
 
 | | The structure of EBOV L-VP35-RNA complex (conformation 1) | | Descriptor: | Polymerase cofactor VP35, RNA-directed RNA polymerase L, The leader sequence of EBOV genome., ... | | Authors: | Qi, P, Yi, S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular mechanism of de novo replication by the Ebola virus polymerase.
Nature, 622, 2023
|
|
6Z58
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003791 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8,14,18,19,22-pentazatetracyclo[13.5.2.12,6.018,21]tricosa-1(21),2,4,6(23),15(22),16,19-heptaen-7-one, SODIUM ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2003791
To Be Published
|
|
6WOT
 
 | | Cryo-EM structure of recombinant rabbit Ryanodine Receptor type 1 mutant R164C in complex with FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
7QGM
 
 | |
7STI
 
 | |
7STJ
 
 | |
7QGL
 
 | |
6ZG0
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
7QGA
 
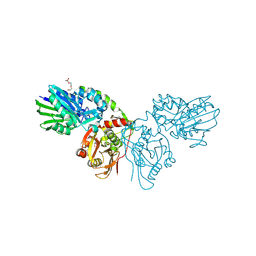 | |
6ZOR
 
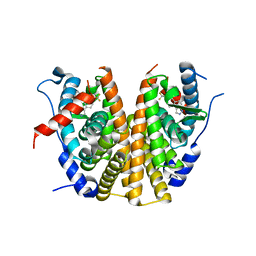 | | Oestrogen receptor ligand binding domain in complex with compound 28 | | Descriptor: | 6-[(6~{S},8~{R})-8-methyl-7-[2,2,2-tris(fluoranyl)ethyl]-3,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-~{N}-(1-propylazetidin-3-yl)pyridin-3-amine, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
7QUW
 
 | | CVB3-3Cpro in complex with inhibitor MG-78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, Protease 3C | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
8FKJ
 
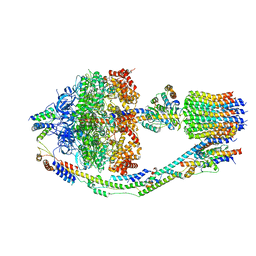 | | Yeast ATP Synthase in conformation-3, at pH 6 | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8JUL
 
 | | Cryo-EM structure of SIDT1 in complex with phosphatidic acid | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, SID1 transmembrane family member 1, ZINC ION | | Authors: | Sun, C.R, Xu, D, Li, Q, Zhou, C.Z, Chen, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Human SIDT1 mediates dsRNA uptake via its phospholipase activity.
Cell Res., 34, 2024
|
|
6WFK
 
 | | Crystal structure of human Naa50 in complex with CoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
7T7Z
 
 | | The crystal structure of family 8 carbohydrate-binding module from Dictyostelium discoideum | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, Endoglucanase, ... | | Authors: | Marcelo, M.V, Campos, B.M, Squina, F.M. | | Deposit date: | 2021-12-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Unique properties of a Dictyostelium discoideum carbohydrate-binding module expand our understanding of CBM-ligand interactions.
J.Biol.Chem., 298, 2022
|
|
7PGQ
 
 | | GAP-SecPH region of human neurofibromin isoform 2 in closed conformation. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
8HAW
 
 | | An auto-activation mechanism of plant non-specific phospholipase C | | Descriptor: | CALCIUM ION, GLYCEROL, Non-specific phospholipase C4, ... | | Authors: | Zhao, F, Fan, R.Y, Guan, Z.Y, Guo, L, Yin, P. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the mechanism of phospholipid hydrolysis by plant non-specific phospholipase C.
Nat Commun, 14, 2023
|
|
8JWD
 
 | | Histidine kinase QseE sensor domain of Escherichia coli O157:H7 | | Descriptor: | 1,2-ETHANEDIOL, histidine kinase | | Authors: | Matsumoto, K, Fukuda, Y, Inoue, T. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structures of QseE and QseG: elements of a three-component system from Escherichia coli.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6HPY
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 12 | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]propanoic acid, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
7SWO
 
 | | C98C7 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | C98C7 Fab heavy chain, C98C7 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
9AZW
 
 | | Crystal structure of PDC-88 beta-lactamase in complex with taniborbactam | | Descriptor: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Beta-lactamase | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2024-03-11 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and mechanism of taniborbactam inhibition of the cefepime-hydrolyzing, partial R2-loop deletion Pseudomonas -derived cephalosporinase variant PDC-88.
Antimicrob.Agents Chemother., 69, 2025
|
|
8ULD
 
 | | SARA CoV-2 3C-like protease in complex with GSK3487016A | | Descriptor: | 1,2-ETHANEDIOL, N-[(benzyloxy)carbonyl]-4-fluoro-L-phenylalanyl-N-{(2S,3R)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]-4-[(propan-2-yl)amino]butan-2-yl}-L-leucinamide, Replicase polyprotein 1a | | Authors: | Williams, S.P, Concha, N.O. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploration of the P1 residue in 3CL protease inhibitors leading to the discovery of a 2-tetrahydrofuran P1 replacement.
Bioorg.Med.Chem., 100, 2024
|
|
7BY6
 
 | | Plasmodium vivax cytoplasmic Phenylalanyl-tRNA synthetase in complex with BRD1389 | | Descriptor: | (3S,4R,8R,9R,10S)-N-(4-cyclopropyloxyphenyl)-10-(methoxymethyl)-3,4-bis(oxidanyl)-9-[4-(2-phenylethynyl)phenyl]-1,6-diazabicyclo[6.2.0]decane-6-carboxamide, MAGNESIUM ION, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Malhotra, N, Manmohan, S, Harlos, K, Melillo, B, Schreiber, S.L, Manickam, Y, Sharma, S. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural basis of malaria parasite phenylalanine tRNA-synthetase inhibition by bicyclic azetidines.
Nat Commun, 12, 2021
|
|
6WEQ
 
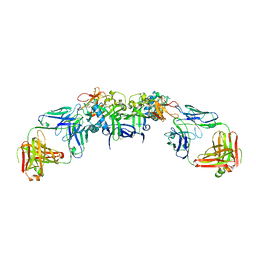 | | DENV1 NS1 in complex with neutralizing 2B7 Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2B7 Fab fragment heavy chain, 2B7 Fab fragment light chain, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2020-04-02 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction.
Science, 371, 2021
|
|
8K5W
 
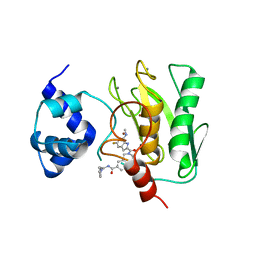 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 2-[[5-fluoranyl-7-(methylamino)-1H-indol-2-yl]carbonyl]-N-(2-pyrrol-1-ylethyl)-3,4-dihydro-1H-isoquinoline-7-carboxamide, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
