4AAU
 
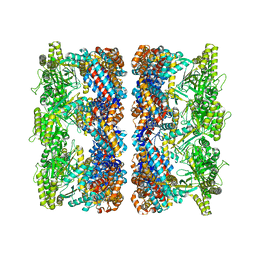 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
3X0C
 
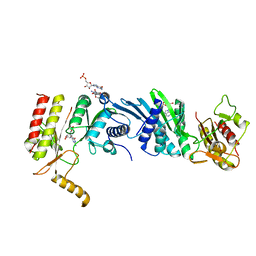 | | Crystal structure of PIP4KIIBETA I368A complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
8HLD
 
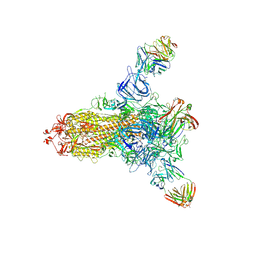 | | S protein of SARS-CoV-2 in complex with 26434 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, Y.Y, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2022-11-29 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A SARS-CoV-2 Spike N-terminal domain neutralizing antibody targets on a glycans shielded silent face and inhibits virus entry via hindering the recognition of RBD and hACE2
To Be Published
|
|
3X08
 
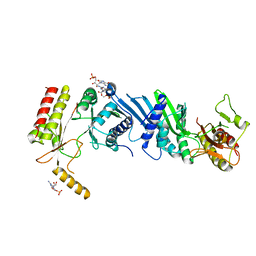 | | Crystal structure of PIP4KIIBETA N203A complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3KIN
 
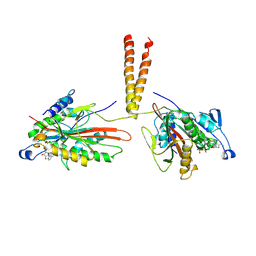 | | KINESIN (DIMERIC) FROM RATTUS NORVEGICUS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN HEAVY CHAIN | | Authors: | Kozielski, F, Sack, S, Marx, A, Thormahlen, M, Schonbrunn, E, Biou, V, Thompson, A, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 1997-08-25 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of dimeric kinesin and implications for microtubule-dependent motility.
Cell(Cambridge,Mass.), 91, 1997
|
|
2OUM
 
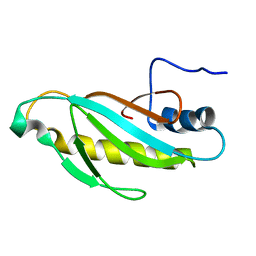 | | The first domain of L1 from Thermus thermophilus | | Descriptor: | 50S ribosomal protein L1 | | Authors: | Kljashtorny, V, Tishchenko, S, Nevskaya, N, Nikonov, S, Davydova, N, Garber, M. | | Deposit date: | 2007-02-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Domain I of ribosomal protein L1 is sufficient for specific RNA binding.
Nucleic Acids Res., 35, 2007
|
|
3X3Q
 
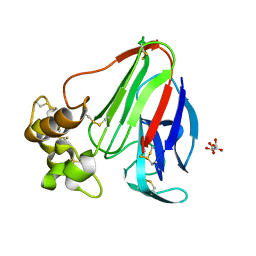 | |
3X07
 
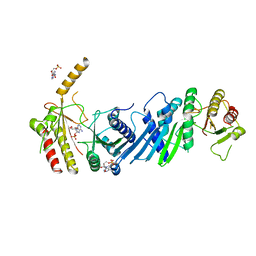 | | Crystal structure of PIP4KIIBETA N203A complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
4AHL
 
 | | P112L - Angiogenin mutants and amyotrophic lateral sclerosis - a biochemical and biological analysis | | Descriptor: | ANGIOGENIN, L(+)-TARTARIC ACID | | Authors: | Thiyagarajan, N, Ferguson, R, Saha, S, Pham, T, Subramanian, V, Acharya, K.R. | | Deposit date: | 2012-02-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structural and Molecular Insights Into the Mechanism of Action of Human Angiogenin-Als Variants in Neurons.
Nat.Commun., 3, 2012
|
|
2P8G
 
 | |
2ORW
 
 | | Thermotoga maritima thymidine kinase 1 like enzyme in complex with TP4A | | Descriptor: | MAGNESIUM ION, P1-(5'-ADENOSYL)P4-(5'-(2'-DEOXY-THYMIDYL))TETRAPHOSPHATE, Thymidine kinase, ... | | Authors: | Segura-Pena, D, Lutz, S, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2007-02-04 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of ATP to TK1-like Enzymes Is Associated with a Conformational Change in the Quaternary Structure.
J.Mol.Biol., 369, 2007
|
|
3WZZ
 
 | | Crystal structure of PIP4KIIBETA | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0U
 
 | | Crystal structure of PirB | | Descriptor: | Uncharacterized protein | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J, Lo, C.F. | | Deposit date: | 2014-10-22 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2OTT
 
 | | Crystal structure of CD5_DIII | | Descriptor: | T-cell surface glycoprotein CD5 | | Authors: | Rodamilans, B. | | Deposit date: | 2007-02-09 | | Release date: | 2007-03-13 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the third extracellular domain of CD5 reveals the fold of a group B scavenger cysteine-rich receptor domain.
J.Biol.Chem., 282, 2007
|
|
3X3R
 
 | |
2OU9
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R1/R119A | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
3KG5
 
 | | Crystal structure of human Ig-beta homodimer | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-28 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
8IG0
 
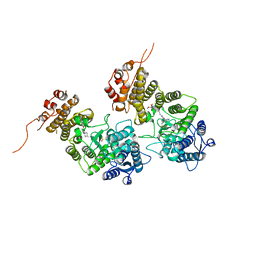 | | Crystal structure of menin in complex with DS-1594b | | Descriptor: | (1R,2S,4R)-4-[[4-(5,6-dimethoxypyridazin-3-yl)phenyl]methylamino]-2-[methyl-[6-[2,2,2-tris(fluoranyl)ethyl]thieno[2,3-d]pyrimidin-4-yl]amino]cyclopentan-1-ol, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Suzuki, M, Yoneyama, T, Imai, E. | | Deposit date: | 2023-02-20 | | Release date: | 2023-03-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel Menin-MLL1 inhibitor, DS-1594a, prevents the progression of acute leukemia with rearranged MLL1 or mutated NPM1.
Cancer Cell Int, 23, 2023
|
|
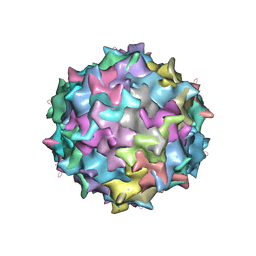
3KIC
 
 | |
3KDM
 
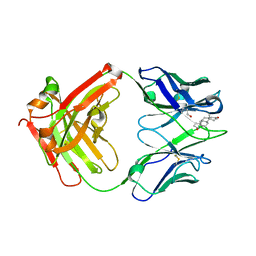 | |
3KUM
 
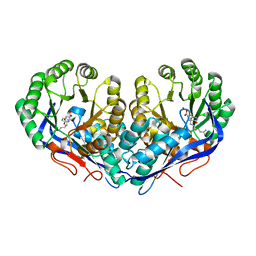 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Arg-L-Tyr | | Descriptor: | ARGININE, Dipeptide Epimerase, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-11-27 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3X3S
 
 | |
2P35
 
 | | Crystal structure of trans-aconitate methyltransferase from Agrobacterium tumefaciens | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Trans-aconitate 2-methyltransferase | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of trans-aconitate methyltransferase from Agrobacterium tumefaciens
To be Published
|
|
2P7P
 
 | | Crystal structure of genomically encoded fosfomycin resistance protein, FosX, from Listeria monocytogenes complexed with MN(II) and sulfate ion | | Descriptor: | Glyoxalase family protein, MANGANESE (II) ION, SULFATE ION | | Authors: | Fillgrove, K.L, Pakhomova, S, Schaab, M, Newcomer, M.E, Armstrong, R.N. | | Deposit date: | 2007-03-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Mechanism of the Genomically Encoded Fosfomycin Resistance Protein, FosX, from Listeria monocytogenes.
Biochemistry, 46, 2007
|
|
3ZL2
 
 | | A thiazolyl-mannoside bound to FimH, orthorhombic space group | | Descriptor: | N-{5-[(1R)-1-hydroxyethyl]-1,3-thiazol-2-yl}-alpha-D-mannopyranosylamine, PROTEIN FIMH | | Authors: | Brument, S, Sivignon, A, Dumych, T.I, Moreau, N, Roos, G, Guerardel, Y, Chalopin, T, Deniaud, D, Bilyy, R.O, Darfeuille-Michaud, A, Bouckaert, J, Gouin, S.G. | | Deposit date: | 2013-01-27 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Thiazolylaminomannosides as Potent Antiadhesives of Type 1 Piliated Escherichia Coli Isolated from Crohn'S Disease Patients.
J.Med.Chem., 56, 2013
|
|
