5OMB
 
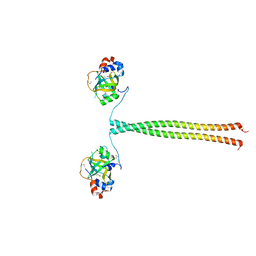 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 N-OB domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA damage checkpoint protein LCD1, ... | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
1XO7
 
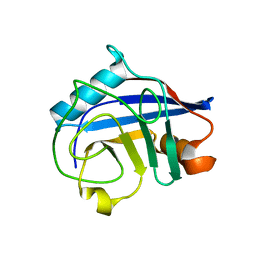 | |
5OMD
 
 | | Crystal structure of S. cerevisiae Ddc2 N-terminal coiled-coil domain | | Descriptor: | DNA damage checkpoint protein LCD1 | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
1GPZ
 
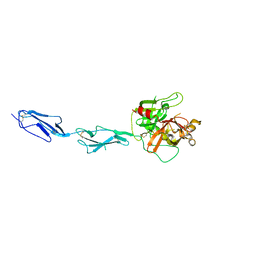 | | THE CRYSTAL STRUCTURE OF THE ZYMOGEN CATALYTIC DOMAIN OF COMPLEMENT PROTEASE C1R | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C1R COMPONENT, ... | | Authors: | Budayova-Spano, M, Fontecilla-Camps, J.C, Gaboriaud, C. | | Deposit date: | 2001-11-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structure of the Zymogen Catalytic Domain of Complement Protease C1R Reveals that a Disruptive Mechanical Stress is Required to Trigger Activation of the C1 Complex.
Embo J., 21, 2002
|
|
3F9R
 
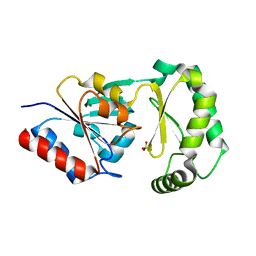 | | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370 | | Descriptor: | MAGNESIUM ION, Phosphomannomutase, SULFATE ION | | Authors: | Wernimont, A.K, Lam, A, Ali, A, Lin, Y.H, Guther, L, Shamshad, A, MacKenzie, F, Bandini, G, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-14 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei phosphomannosemutase, TB.10.700.370
To be Published
|
|
1SM7
 
 | | Solution structure of the recombinant pronapin precursor, BnIb. | | Descriptor: | recombinant Ib pronapin | | Authors: | Pantoja-Uceda, D, Palomares, O, Bruix, M, Villalba, M, Rodriguez, R, Rico, M, Santoro, J. | | Deposit date: | 2004-03-08 | | Release date: | 2005-02-01 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.
Biochemistry, 43, 2004
|
|
5OMC
 
 | | Crystal structure of K. lactis Ddc2 N-terminus in complex with S. cerevisiae Rfa1 (K45E mutant) N-OB domain | | Descriptor: | CHLORIDE ION, DNA damage checkpoint protein LCD1, Replication factor A protein 1 | | Authors: | Deshpande, I, Seeber, A, Shimada, K, Keusch, J.J, Gut, H, Gasser, S.M. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage.
Mol. Cell, 68, 2017
|
|
1XWR
 
 | | Crystal structure of the coliphage lambda transcription activator protein CII | | Descriptor: | ISOPROPYL ALCOHOL, Regulatory protein CII | | Authors: | Datta, A.B, Panjikar, S, Weiss, M.S, Chakrabarti, P, Parrack, P. | | Deposit date: | 2004-11-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure of {lambda} CII: Implications for recognition of direct-repeat DNA by an unusual tetrameric organization
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3FB3
 
 | | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886 | | Descriptor: | N-acetyltransferase | | Authors: | Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, MacKenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-18 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Trypanosoma Brucei Acetyltransferase, Tb11.01.2886
TO BE PUBLISHED
|
|
3Q6J
 
 | | Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme M Oxidoreductase/Carboxylase | | Descriptor: | (2-[2-KETOPROPYLTHIO]ETHANESULFONATE, 1-THIOETHANESULFONIC ACID, 2-oxopropyl-CoM reductase, ... | | Authors: | Pandey, A.S, Mulder, D.W, Ensign, S.A, Peters, J.W. | | Deposit date: | 2011-01-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for carbon dioxide binding by 2-ketopropyl coenzyme M oxidoreductase/carboxylase.
Febs Lett., 585, 2011
|
|
6JKQ
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi (Ligand-free form) | | Descriptor: | Aspartate carbamoyltransferase | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JL6
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with phosphate (Pi). | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, PHOSPHATE ION | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
5ZOR
 
 | |
3H79
 
 | | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70 | | Descriptor: | THIOCYANATE ION, Thioredoxin-like protein | | Authors: | Santos, C.R, Fessel, M.R, Vieira, L.C, Krieger, M.A, Goldenberg, S, Guimaraes, B.G, Zanchin, N.I.T, Barbosa, J.A.R.G. | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70
TO BE PUBLISHED
|
|
3RVA
 
 | | Crystal structure of organophosphorus acid anhydrolase from Alteromonas macleodii | | Descriptor: | MANGANESE (II) ION, NICKEL (II) ION, Organophosphorus acid anhydrolase, ... | | Authors: | Stepankova, A, Koval, T, Ostergaard, L.H, Duskova, J, Skalova, T, Hasek, J, Dohnalek, J. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Organophosphorus acid anhydrolase from Alteromonas macleodii: structural study and functional relationship to prolidases.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1WC7
 
 | |
3UGE
 
 | | Silver Metallated Pseudomonas aeruginosa Azurin at 1.70 A | | Descriptor: | Azurin, SILVER ION | | Authors: | Panzner, M.J, Billinovich, S.M, Parker, J.A, Bladholm, E, Berry, S.M, Ziegler, C.J, Leeper, T.C. | | Deposit date: | 2011-11-02 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Silver Metallation of Pseudomonas aeruginosa Azurin
To be Published
|
|
3UTD
 
 | | Ec_IspH in complex with 4-oxopentyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, 4-oxopentyl trihydrogen diphosphate, FE3-S4 CLUSTER | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-25 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3UV7
 
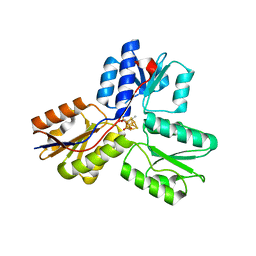 | | Ec_IspH in complex with buta-2,3-dienyl diphosphate (1300) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, buta-2,3-dien-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3UV3
 
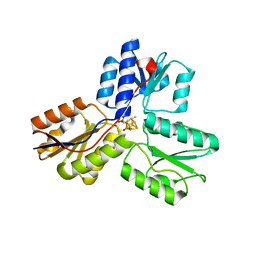 | | Ec_IspH in complex with but-2-ynyl diphosphate (1086) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, but-2-yn-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
1WCB
 
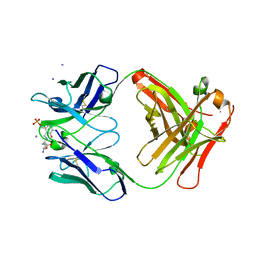 | |
3URK
 
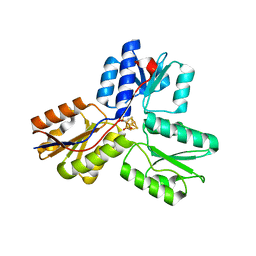 | | IspH in complex with propynyl diphosphate (1061) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, prop-2-yn-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-22 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3UTC
 
 | | Ec_IspH in complex with (E)-4-hydroxybut-3-enyl diphosphate | | Descriptor: | (3E)-4-hydroxybut-3-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Li, K, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-25 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
5SMN
 
 | | PanDDA analysis group deposition of SARS-CoV-2 main protease ligands identified from single sequence-guideddeep generative framework -- Crystal structure of SARS-CoV-2 main protease in complex with Z1365651030 (Mpro-IBM0078) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(1-cyanocyclopropyl)-1-(3-methylpyridin-4-yl)piperidine-4-carboxamide | | Authors: | Fearon, D, Owen, C.D, Lukacik, P, Strain-Damerell, C.M, von Delft, F. | | Deposit date: | 2022-04-20 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | PanDDA analysis group deposition of SARS-CoV-2 main protease ligands identified from single sequence-guideddeep generative framework
To Be Published
|
|
5SML
 
 | | PanDDA analysis group deposition of SARS-CoV-2 main protease ligands identified from single sequence-guideddeep generative framework -- Crystal structure of SARS-CoV-2 main protease in complex with Z68337194 (Mpro-IBM0045) | | Descriptor: | 3C-like proteinase, 6-{[(3,4-dichlorophenyl)methyl](methyl)amino}pyridine-3-sulfonamide, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Lukacik, P, Strain-Damerell, C.M, von Delft, F. | | Deposit date: | 2022-04-20 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition of SARS-CoV-2 main protease ligands identified from single sequence-guideddeep generative framework
To Be Published
|
|
