8AMO
 
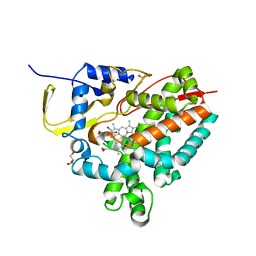 | | Crystal structure of M. tuberculosis CYP143 | | Descriptor: | CHLORIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Grudo, A, Marin, E, Kapranov, I, Shevtsov, M, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
3INM
 
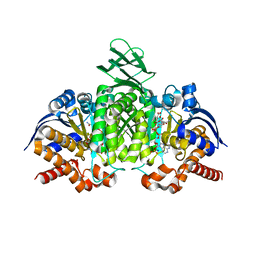 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase R132H mutant in complex with NADPH, ALPHA-KETOGLUTARATE and CALCIUM(2+) | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Fontano, E, Brown, R.S, Suto, R.K, Bhyravbhatla, B. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cancer-associated IDH1 mutations produce 2-hydroxyglutarate.
Nature, 462, 2009
|
|
8AMQ
 
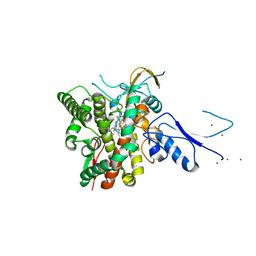 | | Crystal structure of the complex CYP143-FdxE from M. tuberculosis | | Descriptor: | FE3-S4 CLUSTER, NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bukhdruker, S, Varaksa, T, Smolskaya, S, Marin, E, Kapranov, I, Kovalev, K, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
8B0Q
 
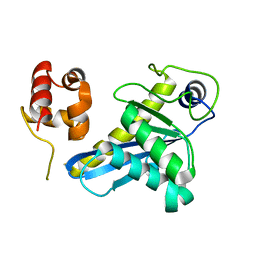 | | Deinococcus radiodurans UvrC C-terminal half | | Descriptor: | UvrABC system protein C | | Authors: | Timmins, J, Stelter, M. | | Deposit date: | 2022-09-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insights into the activation of the dual incision activity of UvrC, a key player in bacterial NER.
Nucleic Acids Res., 51, 2023
|
|
6DGS
 
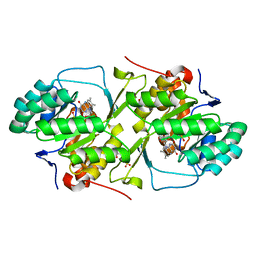 | |
3I94
 
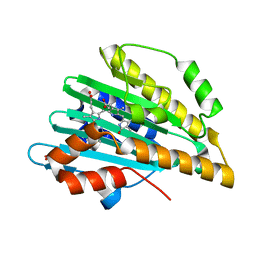 | | Crystal structure of PcyA-biliverdin XIII alpha complex | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-2-ylidene]methy l]-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase, SULFATE ION | | Authors: | Hagiwara, Y, Sugishima, M, Fukuyama, K. | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structural insights into vinyl reduction regiospecificity of phycocyanobilin:ferredoxin oxidoreductase (PcyA).
to be published
|
|
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
2J40
 
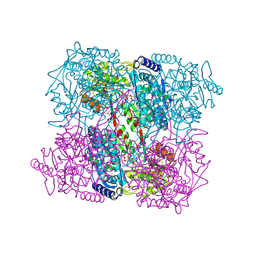 | | 1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound inhibitor L-proline and NAD. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Sakamoto, K, Nishio, M, Yokoyama, S. | | Deposit date: | 2006-08-24 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Ternary Complex of Delta1-Pyrroline-5-Carboxylate Dehydrogenase with Substrate Mimic and Co-Factoer
To be Published
|
|
4EIG
 
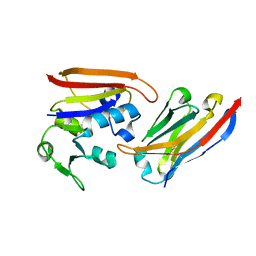 | | CA1698 camel antibody fragment in complex with DHFR | | Descriptor: | CA1698 camel antibody fragment, Dihydrofolate reductase | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
6DPR
 
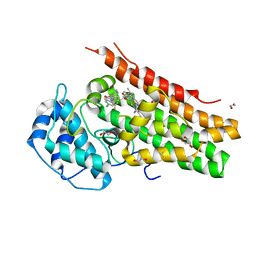 | | Mapping the binding trajectory of a suicide inhibitor in human indoleamine 2,3-dioxygenase 1 | | Descriptor: | (2R)-N-(4-chlorophenyl)-2-[cis-4-(6-fluoroquinolin-4-yl)cyclohexyl]propanamide, 1,2-ETHANEDIOL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Pham, K.N, Yeh, S.R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mapping the Binding Trajectory of a Suicide Inhibitor in Human Indoleamine 2,3-Dioxygenase 1.
J. Am. Chem. Soc., 140, 2018
|
|
2J77
 
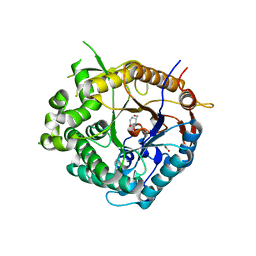 | | Beta-glucosidase from Thermotoga maritima in complex with deoxynojirimycin | | Descriptor: | 1-DEOXYNOJIRIMYCIN, ACETATE ION, BETA-GLUCOSIDASE A, ... | | Authors: | Gloster, T.M, Zechel, D, Davies, G.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
5PAH
 
 | |
3ICE
 
 | | Rho transcription termination factor bound to RNA and ADP-BeF3 | | Descriptor: | 5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Thomsen, N.D, Berger, J.M. | | Deposit date: | 2009-07-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Running in reverse: the structural basis for translocation polarity in hexameric helicases.
Cell(Cambridge,Mass.), 139, 2009
|
|
2WQR
 
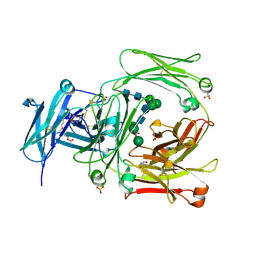 | | The high resolution crystal structure of IgE Fc | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, IG EPSILON CHAIN C REGION, ... | | Authors: | Dhaliwal, B, Sutton, B.J, Beavil, A.J. | | Deposit date: | 2009-08-26 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Changes in Ige Contribute to its Uniquely Slow Dissociation Rate from Receptor Fceri
Nat.Struct.Mol.Biol., 18, 2011
|
|
2J5N
 
 | | 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE FROM THERMUS THERMOPHIRUS WITH BOUND INHIBITOR GLYCINE AND NAD. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Sakamoto, K, Nishio, M, Yokoyama, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of Ternary Complex of Delta1-Pyrroline-5-Carboxylate Dehydrogenase with Substrate Mimic and Co-Factoer
To be Published
|
|
5OUN
 
 | | NMR solution structure of the external DII domain of Rvb2 from Saccharomyces cerevisiae | | Descriptor: | RuvB-like protein 2 | | Authors: | Rouillon, C, Bragantini, B, Charpentier, B, Manival, X, Quinternet, M. | | Deposit date: | 2017-08-24 | | Release date: | 2018-03-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR assignment and solution structure of the external DII domain of the yeast Rvb2 protein.
Biomol NMR Assign, 12, 2018
|
|
6DRV
 
 | |
2J7B
 
 | | Beta-glucosidase from Thermotoga maritima in complex with gluco- tetrazole | | Descriptor: | ACETATE ION, BETA-GLUCOSIDASE A, CALCIUM ION, ... | | Authors: | Gloster, T.M, Zechel, D, Vasella, A, Davies, G.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
8AJJ
 
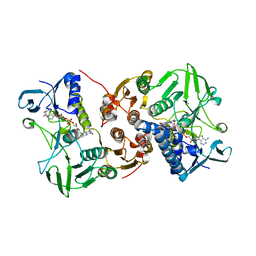 | |
2J4G
 
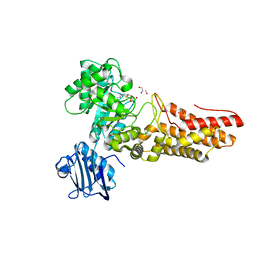 | | Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with n-butyl- thiazoline inhibitor | | Descriptor: | (3AR,5R,6S,7R,7AR)-5-(HYDROXYMETHYL)-2-PROPYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Dennis, R.J, Davies, G.J. | | Deposit date: | 2006-08-31 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of Pugnac and Nag-Thiazoline as Transition State Analogues for Human O-Glcnacase: Mechanistic and Structural Insights Into Inhibitor Selectivity and Transition State Poise.
J.Am.Chem.Soc., 129, 2007
|
|
6YEH
 
 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in apo form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glutamate dehydrogenase 1, POTASSIUM ION | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
2J4S
 
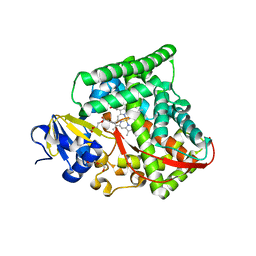 | | P450 BM3 heme domain in complex with DMSO | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kuper, J, Tuck-Seng, W, Roccatano, D, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2006-09-05 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding a Mechanism of Organic Cosolvent Inactivation in Heme Monooxygenase P450 Bm-3.
J.Am.Chem.Soc., 129, 2007
|
|
3IJ7
 
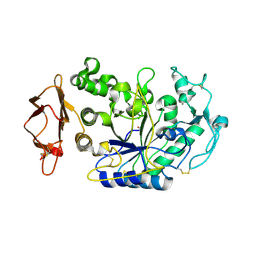 | | Directed 'in situ' Elongation as a Strategy to Characterize the Covalent Glycosyl-Enzyme Catalytic Intermediate of Human Pancreatic a-Amylase | | Descriptor: | 4-O-methyl-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranosyl fluoride, 4-O-methyl-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-2)-5-fluoro-alpha-L-idopyranose, CALCIUM ION, ... | | Authors: | Li, C, Zhang, R, Withers, S.G, Brayer, G.D. | | Deposit date: | 2009-08-03 | | Release date: | 2009-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed "in situ" inhibitor elongation as a strategy to structurally characterize the covalent glycosyl-enzyme intermediate of human pancreatic alpha-amylase
Biochemistry, 48, 2009
|
|
3IJP
 
 | |
2IXA
 
 | | A-zyme, N-acetylgalactosaminidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-N-ACETYLGALACTOSAMINIDASE, ... | | Authors: | Sulzenbacher, G, Liu, Q.P, Bourne, Y, Henrissat, B, Clausen, H. | | Deposit date: | 2006-07-07 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial Glycosidases for the Production of Universal Red Blood Cells.
Nat.Biotechnol., 25, 2007
|
|
