2YGH
 
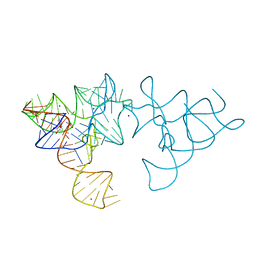 | | SAM-I riboswitch with a G2nA mutation in the Kink turn in complex with S-adenosylmethionine | | Descriptor: | POTASSIUM ION, S-ADENOSYLMETHIONINE, SAM-I RIBOSWITCH, ... | | Authors: | Schroeder, K, Daldrop, P, Lilley, D. | | Deposit date: | 2011-04-17 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RNA Tertiary Interactions in a Riboswitch Stabilize the Structure of a Kink Turn.
Structure, 19, 2011
|
|
4EML
 
 | |
8OKB
 
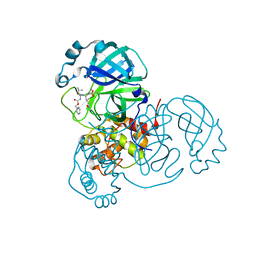 | | SARS-CoV2 NSP5 in complex with a peptidomimetic ligand | | Descriptor: | 3C-like proteinase nsp5, methyl (4~{S})-4-[[(2~{S})-4-methyl-2-(phenylmethoxycarbonylamino)pentanoyl]amino]-5-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Calderone, V. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
8OKC
 
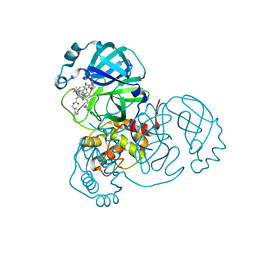 | | SARS-CoV2 NSP5 in complex with a GC-376 based peptidomimetic PROTAC | | Descriptor: | (phenylmethyl) ~{N}-[(2~{R})-1-[[(~{Z},2~{S})-5-[4-[[1-[2-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]-6-fluoranyl-1,3-bis(oxidanylidene)isoindol-5-yl]piperidin-4-yl]methyl]piperazin-1-yl]-5-oxidanylidene-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]pent-3-en-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Calderone, V. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
2Z41
 
 | | Crystal Structure Analysis of the Ski2-type RNA helicase | | Descriptor: | MAGNESIUM ION, putative ski2-type helicase | | Authors: | Nakashima, T, Zhang, X, Kakuta, Y, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2007-06-12 | | Release date: | 2008-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Crystal structure of an archaeal Ski2p-like protein from Pyrococcus horikoshii OT3
Protein Sci., 17, 2008
|
|
2JI3
 
 | | X-ray structure of the iron-peroxide intermediate of superoxide reductase (E114A mutant) from Desulfoarculus baarsii | | Descriptor: | CALCIUM ION, Desulfoferrodoxin, FE (III) ION, ... | | Authors: | Katona, G, Carpentier, P, Niviere, V, Amara, P, Adam, V, Ohana, J, Tsanov, N, Bourgeois, D. | | Deposit date: | 2007-02-24 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme.
Science, 316, 2007
|
|
5I5Z
 
 | | CDK8-CYCC IN COMPLEX WITH 8-(1-Methyl-2,2-dioxo-2,3-dihydro-1H-2l6-benzo[c]isothiazol-5-yl)-[1,6]naphthyridine-2-carboxylic acid methylamide | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, FORMIC ACID, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2016-02-15 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2,8-Disubstituted-1,6-Naphthyridines and 4,6-Disubstituted-Isoquinolines with Potent, Selective Affinity for CDK8/19.
Acs Med.Chem.Lett., 7, 2016
|
|
5I86
 
 | | Crystal structure of the bromodomain of human CREBBP bound to the benzodiazepinone G02778174 | | Descriptor: | (4R)-N-benzyl-4-methyl-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepine-6-carboxamide, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5IS5
 
 | |
4Y30
 
 | | Crystal structure of human protein arginine methyltransferase PRMT6 bound to SAH and EPZ020411 | | Descriptor: | GLYCEROL, MAGNESIUM ION, N,N'-dimethyl-N-({3-[4-({trans-3-[2-(tetrahydro-2H-pyran-4-yl)ethoxy]cyclobutyl}oxy)phenyl]-1H-pyrazol-4-yl}methyl)ethane-1,2-diamine, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound.
Acs Med.Chem.Lett., 6, 2015
|
|
2D5K
 
 | | Crystal structure of Dps from Staphylococcus aureus | | Descriptor: | Dps family protein, GLYCEROL | | Authors: | Tanaka, Y, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleoid compaction by MrgA(Asp56Ala/Glu60Ala) does not contribute to staphylococcal cell survival against oxidative stress and phagocytic killing by macrophages
FEMS Microbiol. Lett., 360, 2014
|
|
5IVT
 
 | | Crystal Structure of HIV Protease complexed with [(1S)-1-[(S)-(4-chlorophenyl)-(3,5-difluorophenyl)methyl]-2-[[5-fluoro-4-[2-[(2R,5S)-5-(2,2,2-trifluoroethylcarbamoyloxymethyl)morpholin-4-ium-2-yl]ethyl]pyridin-1-ium-3-yl]amino]-2-oxo-ethyl]ammonium | | Descriptor: | (betaS)-4-chloro-beta-(3,5-difluorophenyl)-N-(5-fluoro-4-{2-[(2R,5S)-5-({[(2,2,2-trifluoroethyl)carbamoyl]oxy}methyl)morpholin-2-yl]ethyl}pyridin-3-yl)-L-phenylalaninamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Su, H.P. | | Deposit date: | 2016-03-21 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery of MK-8718, an HIV Protease Inhibitor Containing a Novel Morpholine Aspartate Binding Group.
Acs Med.Chem.Lett., 7, 2016
|
|
5LVR
 
 | | Crystal structure of human PCAF bromodomain in complex with compound-E (CPD-E) | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-2-phenyl-1,2,3-triazole-4-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hopkins, A.L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
5J6S
 
 | | Crystal structure of Endoplasmic Reticulum Aminopeptidase 2 (ERAP2) in complex with a hydroxamic derivative ligand | | Descriptor: | (2S)-N~1~-benzyl-2-[(4-fluorophenyl)methyl]-N~3~-hydroxypropanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saridakis, E, Giastas, P, Mpakali, A, Deprez-Poulain, R, Stratikos, E. | | Deposit date: | 2016-04-05 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
5IXY
 
 | | Lactate Dehydrogenase in complex with hydroxylactam inhibitor compound 31: (2~{S})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one | | Descriptor: | (2~{S})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one, L-lactate dehydrogenase A chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chen, Z, Eigenbrot, C. | | Deposit date: | 2016-03-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice.
Acs Med.Chem.Lett., 7, 2016
|
|
5LVQ
 
 | | Crystal structure of human PCAF bromodomain in complex with compound-D (CPD-D), N-methyl-2-(tetrahydro-2H-pyran-4-yloxy)benzamide | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Histone acetyltransferase KAT2B, ... | | Authors: | Chaikuad, A, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hopkins, A.L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of New Bromodomain Scaffolds by Biosensor Fragment Screening.
ACS Med Chem Lett, 7, 2016
|
|
5IXS
 
 | | Lactate Dehydrogenase in complex with hydroxylactam inhibitor compound 9: (6R)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(3-hydroxyphenyl)-6-(thiophen-3-yl)-5,6-dihydropyridin-2(1H)-one | | Descriptor: | (6R)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(3-hydroxyphenyl)-6-(thiophen-3-yl)-5,6-dihydropyridin-2(1H)-one, 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2016-03-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice.
Acs Med.Chem.Lett., 7, 2016
|
|
1FFN
 
 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH PEPTIDE GP33(C9M) | | Descriptor: | BETA-2 MICROGLOBULIN, BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition
J.IMMUNOL., 169, 2002
|
|
5M6U
 
 | | HUMAN PI3KDELTA IN COMPLEX WITH LASW1579 | | Descriptor: | 4-azanyl-6-[[(1~{S})-1-(4-oxidanylidene-3-phenyl-pyrrolo[2,1-f][1,2,4]triazin-2-yl)ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Segarra, V, Hernandez, B, Lozoya, E, Blaesse, M, Hoeppner, S, Jestel, A. | | Deposit date: | 2016-10-26 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a Potent, Selective, and Orally Available PI3K delta Inhibitor for the Treatment of Inflammatory Diseases.
ACS Med Chem Lett, 8, 2017
|
|
5K9D
 
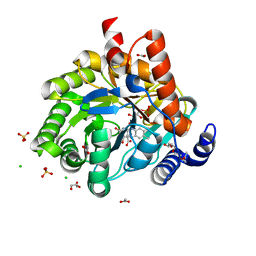 | | Crystal structure of human dihydroorotate dehydrogenase at 1.7 A resolution | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lewis, T.A, Sykes, D.B, Law, J.M, Munoz, B, Scadden, D.T, Rustiguel, J.K, Nonato, M.C, Schreiber, S.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia.
ACS Med Chem Lett, 7, 2016
|
|
5JY3
 
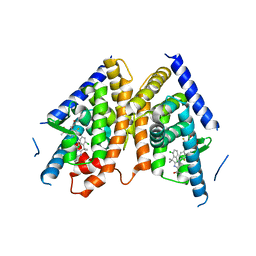 | |
2JI1
 
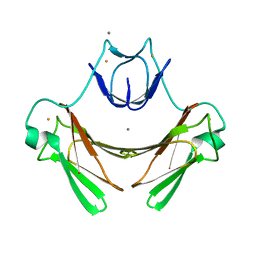 | | X-ray structure of wild-type superoxide reductase from Desulfoarculus baarsii | | Descriptor: | CALCIUM ION, Desulfoferrodoxin, FE (II) ION, ... | | Authors: | Katona, G, Carpentier, P, Niviere, V, Amara, P, Adam, V, Ohana, J, Tsanov, N, Bourgeois, D. | | Deposit date: | 2007-02-24 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme.
Science, 316, 2007
|
|
4XUI
 
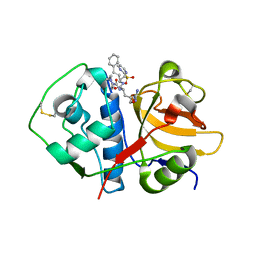 | |
5MPK
 
 | | Crystal structure of CREBBP bromodomain complexed with DK19 | | Descriptor: | CREB-binding protein, ~{N}-(5-ethanoyl-2-ethoxy-phenyl)-3-(2~{H}-1,2,3,4-tetrazol-5-yl)-5-(1,3-thiazol-4-yl)benzamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
2JI2
 
 | | X-ray structure of E114A mutant of superoxide reductase from Desulfoarculus baarsii in the native, reduced form | | Descriptor: | CALCIUM ION, Desulfoferrodoxin, FE (II) ION, ... | | Authors: | Katona, G, Carpentier, P, Niviere, V, Amara, P, Adam, V, Ohana, J, Tsanov, N, Bourgeois, D. | | Deposit date: | 2007-02-24 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme.
Science, 316, 2007
|
|
