6X4L
 
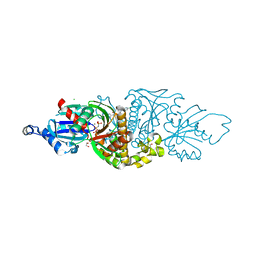 | | PANK3 complex structure with compound PZ-3565 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(5-chloropyrazin-2-yl)piperazin-1-yl]-2-[4-(propan-2-yl)phenyl]ethan-1-one, CHLORIDE ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | LipE guided discovery of isopropylphenyl pyridazines as pantothenate kinase modulators.
Bioorg.Med.Chem., 52, 2021
|
|
3FYA
 
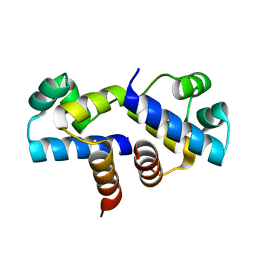 | | Crystal Structure of an R35A mutant of the Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | Regulatory protein | | Authors: | Ball, N.J, McGeehan, J.E, Thresh, S.J, Streeter, S.D, Kneale, G.G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the restriction-modification controller protein C.Esp1396I.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4YG6
 
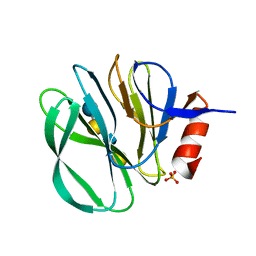 | | Structural basis of glycan recognition in neonate-specific rotaviruses | | Descriptor: | Outer capsid protein VP8*, PHOSPHATE ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, L, Prasad, B.V.V. | | Deposit date: | 2015-02-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural basis of glycan specificity in neonate-specific bovine-human reassortant rotavirus.
Nat Commun, 6, 2015
|
|
6X4K
 
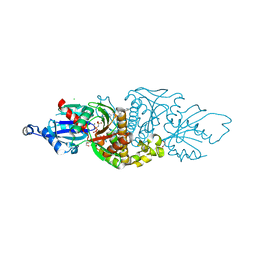 | | PANK3 complex structure with compound PZ-2890 | | Descriptor: | 1,2-ETHANEDIOL, 4-(6-cyanopyridazin-3-yl)-N-[4-(propan-2-yl)phenyl]-3,4-dihydropyrazine-1(2H)-carboxamide, CHLORIDE ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | LipE guided discovery of isopropylphenyl pyridazines as pantothenate kinase modulators.
Bioorg.Med.Chem., 52, 2021
|
|
6X4J
 
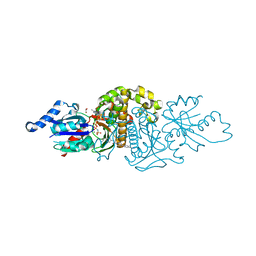 | | PANK3 complex structure with compound PZ-2863 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-{[4-(propan-2-yl)phenyl]acetyl}piperazin-1-yl)pyridine-3-carbonitrile, MAGNESIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | LipE guided discovery of isopropylphenyl pyridazines as pantothenate kinase modulators.
Bioorg.Med.Chem., 52, 2021
|
|
2X4O
 
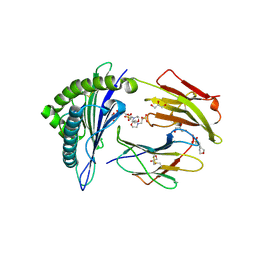 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 envelope peptide env120-128 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, ENVELOPE GLYCOPROTEIN GP160, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2E2M
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (sulfinic acid form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | Deposit date: | 2006-11-14 | | Release date: | 2007-11-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6AOC
 
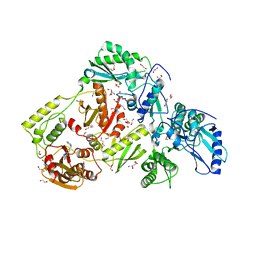 | | Crystal Structure of an N-Hydroxythienopyrimidine-2,4-dione RNase H Active Site Inhibitor with Multiple Binding Modes to HIV Reverse Transcriptase | | Descriptor: | 1,2-ETHANEDIOL, 6-benzyl-3-hydroxythieno[2,3-d]pyrimidine-2,4(1H,3H)-dione, MANGANESE (II) ION, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-08-15 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis and biological evaluations of N-Hydroxy thienopyrimidine-2,4-diones as inhibitors of HIV reverse transcriptase-associated RNase H.
Eur J Med Chem, 141, 2017
|
|
1LLF
 
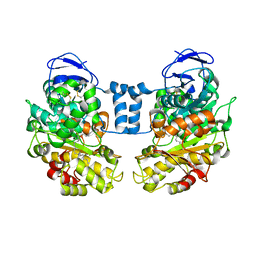 | | Cholesterol Esterase (Candida Cylindracea) Crystal Structure at 1.4A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase 3, TRICOSANOIC ACID | | Authors: | Pletnev, V, Addlagatta, A, Wawrzak, Z, Duax, W. | | Deposit date: | 2002-04-28 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Three-dimensional structure of homodimeric cholesterol esterase-ligand complex at 1.4 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6AUZ
 
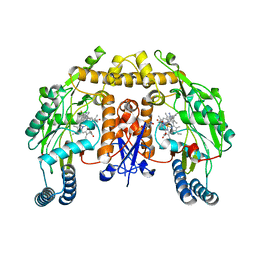 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(3-Fluoro-5-(3-(methylamino)propyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-fluoro-5-[3-(methylamino)propyl]phenyl}ethyl)-4-methylpyridin-2-amine, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
2DZY
 
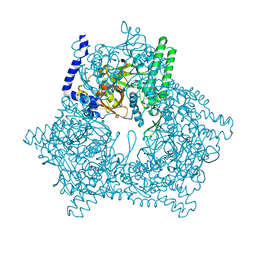 | |
2WM5
 
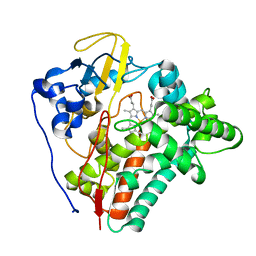 | | X-ray structure of the substrate-free Mycobacterium tuberculosis cytochrome P450 CYP124 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 124 | | Authors: | Johnston, J.B, Kells, P.M, Podust, L.M, Ortiz de Montellano, P.R. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Biochemical and structural characterization of CYP124: a methyl-branched lipid omega-hydroxylase from Mycobacterium tuberculosis.
Proc. Natl. Acad. Sci. U.S.A., 106, 2009
|
|
4Y45
 
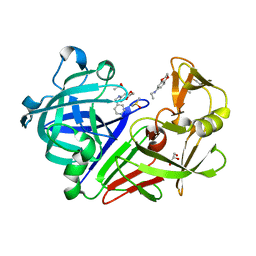 | |
6X7E
 
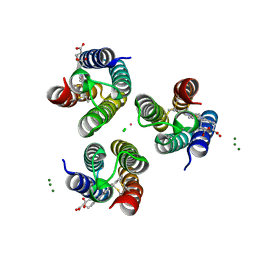 | | Co-bound structure of an engineered protein trimer, TriCyt3, with delta isomerism at the hexahistidine coordination site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, HEME C, ... | | Authors: | Tezcan, F.A, Kakkis, A. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9987 Å) | | Cite: | Metal-Templated Design of Chemically Switchable Protein Assemblies with High-Affinity Coordination Sites.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1LNS
 
 | |
4Y4H
 
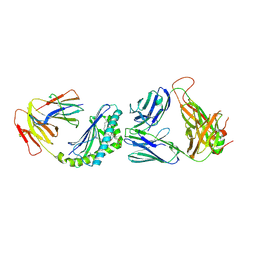 | | Crystal structure of the mCD1d/GCK152/iNKTCR ternary complex | | Descriptor: | (1R)-1,5-anhydro-1-{(1E,3S,4S,5R)-4,5-dihydroxy-3-[(8-phenyloctanoyl)amino]nonadec-1-en-1-yl}-D-galactitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Yu, E.D. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural modifications of alphaGalCer in both lipid and carbohydrate moiety influence activation of murine and human iNKT cells
To Be Published
|
|
2WP3
 
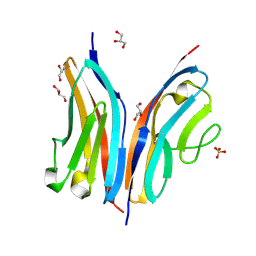 | | Crystal structure of the Titin M10-Obscurin like 1 Ig complex | | Descriptor: | GLYCEROL, OBSCURIN-LIKE PROTEIN 1, SULFATE ION, ... | | Authors: | Pernigo, S, Fuzukawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2009-08-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Insight Into M-Band Assembly and Mechanics from the Titin-Obscurin-Like-1 Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2E5D
 
 | | Crystal structure of Human NMPRTase complexed with nicotinamide | | Descriptor: | NICOTINAMIDE, Nicotinamide phosphoribosyltransferase | | Authors: | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
2E5U
 
 | |
6AW8
 
 | | 2.25A resolution domain swapped dimer structure of SAH bound catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
4Y5S
 
 | | Structure of FtmOx1 with a-Ketoglutarate as co-substrate | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, FE (II) ION, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
4IPP
 
 | |
6X80
 
 | | Structure of the Campylobacter jejuni G508A Flagellar Filament | | Descriptor: | 5,7-diamino-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid, Flagellin A | | Authors: | Kreutzberger, M.A.B, Wang, F, Egelman, E.H. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structure of the Campylobacter jejuni flagellar filament reveals how epsilon Proteobacteria escaped Toll-like receptor 5 surveillance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6AN3
 
 | | Crystal structure of H172A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant soaked with peptide (no CuH bound, no peptide bound) | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AUB
 
 | |
