3C6V
 
 | | Crystal structure of AU4130/APC7354, a probable enzyme from the thermophilic fungus Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, Probable tautomerase/dehalogenase AU4130, SODIUM ION, ... | | Authors: | Singer, A.U, Binkowski, T.A, Skarina, T, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-05 | | Release date: | 2008-02-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of AU4130/APC7354, a probable enzyme from the thermophilic fungus Aspergillus fumigatus.
To be Published
|
|
3M1A
 
 | | The Crystal Structure of a Short-chain Dehydrogenase from Streptomyces avermitilis to 2A | | Descriptor: | ACETATE ION, Putative dehydrogenase, SODIUM ION | | Authors: | Stein, A.J, Evdokimova, E, Egorova, O, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-03-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of a Short-chain Dehydrogenase from Streptomyces avermitilis to 2A
To be Published
|
|
3LZK
 
 | | The crystal structure of a probably aromatic amino acid degradation proteiN from Sinorhizobium meliloti 1021 | | Descriptor: | CALCIUM ION, Fumarylacetoacetate hydrolase family protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-01 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a probably aromatic amino acid degradation protein from Sinorhizobium meliloti 1021
To be Published
|
|
3M8A
 
 | | Crystal Structure of Swine Flu Virus NS1 N-Terminal RNA Binding Domain from H1N1 Influenza A/California/07/2009 | | Descriptor: | ACETATE ION, MALONATE ION, Nonstructural protein 1, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Swine Flu Virus NS1 N-Terminal RNA Binding Domain from H1N1 Influenza A/California/07/2009
To be Published
|
|
4LAT
 
 | | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A in complex with phosphate | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHATE ION, Phosphate-binding protein PstS 1 | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A in complex with phosphate
To be Published
|
|
3CZQ
 
 | | Crystal structure of putative polyphosphate kinase 2 from Sinorhizobium meliloti | | Descriptor: | FORMIC ACID, GLYCEROL, Putative polyphosphate kinase 2 | | Authors: | Osipiuk, J, Evdokimova, E, Nocek, B, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3KC2
 
 | | Crystal structure of mitochondrial HAD-like phosphatase from Saccharomyces cerevisiae | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Uncharacterized protein YKR070W | | Authors: | Nocek, B, Evdokimova, E, Kuznetsova, K, Iakunine, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of mitochondrial HAD-like phosphatase from Saccharomyces cerevisiae
To be Published
|
|
3MAJ
 
 | | Crystal structure of putative DNA processing protein DprA from Rhodopseudomonas palustris CGA009 | | Descriptor: | DNA processing chain A, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-23 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of putative DNA processing protein DprA from Rhodopseudomonas palustris
To be Published
|
|
4JCC
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiuA from Streptococcus pneumoniae Canada MDR_19A | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, Iron-compound ABC transporter, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-21 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.133 Å) | | Cite: | Crystal structure of iron uptake ABC transporter substrate-binding protein PiuA from Streptococcus pneumoniae Canada MDR_19A
TO BE PUBLISHED
|
|
7V09
 
 | | Crystal structure of ECL_RS08780, putative sugar transport system periplasmic sugar-binding protein | | Descriptor: | MAGNESIUM ION, Multiple sugar transport system periplasmic sugar-binding protein | | Authors: | Stogios, P.J, Skarina, T, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ECL_RS08780, putative sugar transport system periplasmic sugar-binding protein
To Be Published
|
|
3MZ1
 
 | | The crystal structure of a possible TRANSCRIPTION REGULATOR PROTEIN from Sinorhizobium meliloti 1021 | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator | | Authors: | Tan, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-11 | | Release date: | 2010-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The crystal structure of a possible TRANSCRIPTION REGULATOR PROTEIN from Sinorhizobium meliloti 1021
To be Published
|
|
3MVK
 
 | | The Crystal Structure of FucU from Bifidobacterium longum to 1.65A | | Descriptor: | GLYCEROL, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Stein, A.J, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of FucU from Bifidobacterium longum to 1.65A
To be Published
|
|
3MVP
 
 | | The Crystal Structure of a TetR/AcrR transcriptional regulator from Streptococcus mutans to 1.85A | | Descriptor: | TetR/AcrR transcriptional regulator | | Authors: | Stein, A.J, Xu, X, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of a TetR/AcrR transcriptional regulator from Streptococcus mutans to 1.85A
To be Published
|
|
4JG9
 
 | | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis | | Descriptor: | Lipoprotein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis
To be Published
|
|
3BIH
 
 | | Crystal structure of fructose-1,6-bisphosphatase from E.coli GlpX | | Descriptor: | Fructose-1,6-bisphosphatase class II glpX, UNKNOWN ATOM OR ION | | Authors: | Lunin, V.V, Skarina, T, Brown, G, Yakunin, A.F, Edwards, A.M, Savchenko, A. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3DR8
 
 | | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium with its cofactor Acetyl-CoA | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Singer, A.U, Skarina, T, Onopriyenko, O, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of yncA, a putative ACETYLTRANSFERASE from Salmonella typhimurium with its cofactor Acetyl-CoA
To be Published
|
|
3MTJ
 
 | | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A | | Descriptor: | Homoserine dehydrogenase, SULFATE ION | | Authors: | Stein, A.J, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of a Homoserine Dehydrogenase from Thiobacillus denitrificans to 2.15A
To be Published
|
|
3CQY
 
 | | Crystal structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1 | | Descriptor: | Anhydro-N-acetylmuramic acid kinase, CHLORIDE ION, SUCCINIC ACID | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1.
To be Published
|
|
3KKD
 
 | | Structure of a putative tetr transcriptional regulator (pa3699) from pseudomonas aeruginosa pa01 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Filippova, E.V, Chruszcz, M, Cymborowski, M, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-05 | | Release date: | 2009-12-15 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Putative TetR Transcriptional Regulator (PA3699) from Pseudomonas Aeruginosa PA01
To be Published
|
|
3N99
 
 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, uncharacterized protein TM1086 | | Authors: | Chruszcz, M, Domagalski, M.J, Wang, S, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
3N0L
 
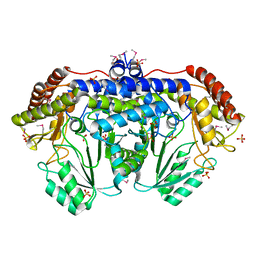 | | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni | | Descriptor: | SULFATE ION, Serine hydroxymethyltransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-14 | | Release date: | 2010-05-26 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of serine hydroxymethyltransferase from Campylobacter jejuni
TO BE PUBLISHED
|
|
3N73
 
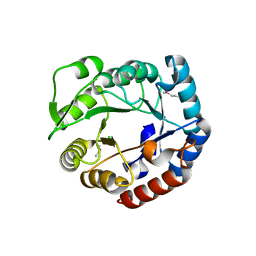 | | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus | | Descriptor: | CHLORIDE ION, Putative 4-hydroxy-2-oxoglutarate aldolase | | Authors: | Cabello, R, Chruszcz, M, Xu, X, Zimmerman, M.D, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus
To be Published
|
|
3NA2
 
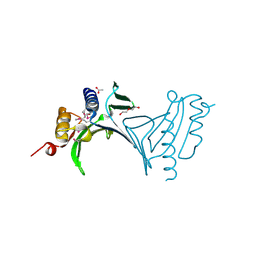 | | Crystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Uncharacterized protein | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crsystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum
To be Published
|
|
3MQZ
 
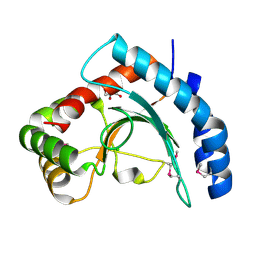 | | Crystal Structure of Conserved Protein DUF1054 from Pink Subaerial Biofilm Microbial Leptospirillum sp. Group II UBA. | | Descriptor: | CHLORIDE ION, GLYCEROL, uncharacterized Conserved Protein DUF1054 | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-28 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Conserved Protein DUF1054 from Pink Subaerial Biofilm Microbial Leptospirillum sp. Group II UBA.
To be Published
|
|
3NI7
 
 | | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718 | | Descriptor: | Bacterial regulatory proteins, TetR family | | Authors: | Knapik, A, Chruszcz, M, Cymborowski, M, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718
To be Published
|
|
