8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | 著者 | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | 登録日 | 2023-05-13 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8JDA
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2 | | 分子名称: | Sodium/hydrogen exchanger 7 | | 著者 | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | 登録日 | 2023-05-13 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
7CMI
 
 | | The LAT2-4F2hc complex in complex with leucine | | 分子名称: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | 著者 | Yan, R.H, Zhou, J.Y, Li, Y.N, Lei, J.L, Zhou, Q. | | 登録日 | 2020-07-27 | | 公開日 | 2020-12-23 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insight into the substrate recognition and transport mechanism of the human LAT2-4F2hc complex.
Cell Discov, 6, 2020
|
|
5CK6
 
 | | Crystal structure of SZ348 in complex with cyclopentene oxide | | 分子名称: | (1R,5S)-6-oxabicyclo[3.1.0]hexane, Limonene-1,2-epoxide hydrolase, NICKEL (II) ION, ... | | 著者 | Wu, L, Sun, Z.T, Reetz, M.T, Zhou, J.H. | | 登録日 | 2015-07-15 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of SZ348 in complex with cyclopentene oxide
To Be Published
|
|
4LJY
 
 | | Crystal structure of RNA splicing effector Prp5 in complex with ADP | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Zhang, Z.-M, Li, J, Yang, F, Xu, Y, Zhou, J. | | 登録日 | 2013-07-05 | | 公開日 | 2013-12-11 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly.
Cell Rep, 5, 2013
|
|
5CLK
 
 | | Crystal structure of SZ348 in complex with S,S-cyclohexanediol | | 分子名称: | (1S,2S)-cyclohexane-1,2-diol, CHLORIDE ION, Limonene-1,2-epoxide hydrolase, ... | | 著者 | Wu, L, Sun, Z.T, Reetz, M.T, Zhou, J.H. | | 登録日 | 2015-07-16 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Crystal structure of SZ348 in complex with S,S-cyclohexanediol
To Be Published
|
|
7V2U
 
 | | The complex structure of SoBcmB and its product 2f | | 分子名称: | (1S,5S,6S)-5-methyl-1-[(1S,2S)-2-methyl-1,2,3-tris(oxidanyl)propyl]-2-oxa-7,9-diazabicyclo[4.2.2]decane-8,10-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | 著者 | Wu, L, Zhou, J.H. | | 登録日 | 2021-08-09 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.00009918 Å) | | 主引用文献 | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2X
 
 | | The complex structure of soBcmB and its substrate 1 | | 分子名称: | (3S,6S)-3-[(2S)-butan-2-yl]-6-[(2R)-2-methyl-2,3-bis(oxidanyl)propyl]piperazine-2,5-dion, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | 著者 | Wu, L, Zhou, J.H. | | 登録日 | 2021-08-10 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.08387423 Å) | | 主引用文献 | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
5CF1
 
 | |
8HP6
 
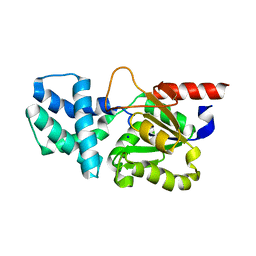 | | Crystal structure of (S)-2-haloacid dehalogenase D12A mutant | | 分子名称: | (S)-2-haloacid dehalogenase, SODIUM ION | | 著者 | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-12-12 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP5
 
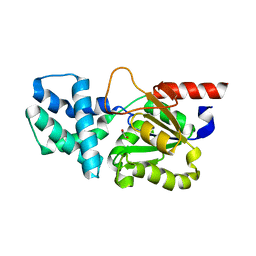 | | Crystal structure of (S)-2-haloacid dehalogenase | | 分子名称: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL | | 著者 | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-12-12 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
4LK2
 
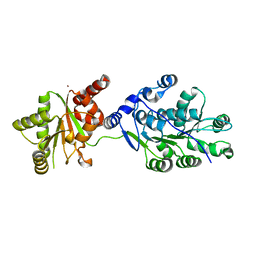 | | Crystal structure of RNA splicing effector Prp5 | | 分子名称: | NICKEL (II) ION, Pre-mRNA-processing ATP-dependent RNA helicase PRP5 | | 著者 | Zhang, Z.-M, Li, J, Yang, F, Xu, Y, Zhou, J. | | 登録日 | 2013-07-05 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly.
Cell Rep, 5, 2013
|
|
2HZ6
 
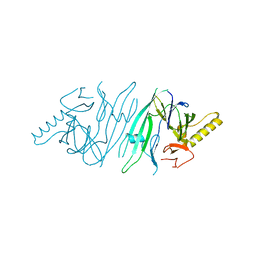 | | The crystal structure of human IRE1-alpha luminal domain | | 分子名称: | Endoplasmic reticulum to nucleus signalling 1 isoform 1 variant | | 著者 | Kaufman, R.J, Xu, Z, Zhou, J. | | 登録日 | 2006-08-08 | | 公開日 | 2006-08-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7V34
 
 | | The complex structure of soBcmB and its product 1d | | 分子名称: | (3S,4S,5S,8S)-8-[(2S)-butan-2-yl]-3-methyl-3,4-bis(oxidanyl)-1-oxa-7,10-diazaspiro[4.5]decane-6,9-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | 著者 | Wu, L, Zhou, J.H. | | 登録日 | 2021-08-10 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.000169 Å) | | 主引用文献 | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V3N
 
 | | The complex structure of soBcmB-D307A and its natural precursor 2 | | 分子名称: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | 著者 | Wu, L, Zhou, J.H. | | 登録日 | 2021-08-10 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.850011 Å) | | 主引用文献 | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7CMH
 
 | | The LAT2-4F2hc complex in complex with tryptophan | | 分子名称: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | 著者 | Yan, R.H, Zhou, J.Y, Li, Y.N, Lei, J.L, Zhou, Q. | | 登録日 | 2020-07-27 | | 公開日 | 2020-12-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural insight into the substrate recognition and transport mechanism of the human LAT2-4F2hc complex.
Cell Discov, 6, 2020
|
|
8WKP
 
 | |
8WQ4
 
 | |
8WQ2
 
 | |
4OU4
 
 | | Crystal structure of esterase rPPE mutant S159A complexed with (S)-Ac-CPA | | 分子名称: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-02-15 | | 公開日 | 2014-07-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OB6
 
 | | Complex structure of esterase rPPE S159A/W187H and substrate (S)-Ac-CPA | | 分子名称: | (2S)-(acetyloxy)(2-chlorophenyl)ethanoic acid, Alpha/beta hydrolase fold-3 domain protein | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Chen, Q, Zhou, J.H, Xu, J.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OB8
 
 | | Crystal structure of a novel thermostable esterase from Pseudomonas putida ECU1011 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OB7
 
 | | Crystal structure of esterase rPPE mutant W187H | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-01-07 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4OU5
 
 | | Crystal structure of esterase rPPE mutant S159A/W187H | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase fold-3 domain protein, DI(HYDROXYETHYL)ETHER | | 著者 | Dou, S, Kong, X.D, Ma, B.D, Xu, J.H, Zhou, J.H. | | 登録日 | 2014-02-15 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal structures of Pseudomonas putida esterase reveal the functional role of residues 187 and 287 in substrate binding and chiral recognition
Biochem.Biophys.Res.Commun., 446, 2014
|
|
4MYS
 
 | | 1.4 Angstrom Crystal Structure of 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase with SHCHC and Pyruvate | | 分子名称: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, GLYCEROL, ... | | 著者 | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | 登録日 | 2013-09-28 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.423 Å) | | 主引用文献 | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
