7BQF
 
 | | Dimerization of SAV1 WW tandem | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, Protein salvador homolog 1 | | 著者 | Lin, Z, Zhang, M. | | 登録日 | 2020-03-24 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.70037615 Å) | | 主引用文献 | A WW Tandem-Mediated Dimerization Mode of SAV1 Essential for Hippo Signaling.
Cell Rep, 32, 2020
|
|
7CDB
 
 | | Structure of GABARAPL1 in complex with GABA(A) receptor gamma 2 | | 分子名称: | ACETATE ION, CITRIC ACID, Gamma-aminobutyric acid receptor subunit gamma-2, ... | | 著者 | Li, J, Ye, J, Zhu, R, Kong, C, Zhang, M, Wang, C. | | 登録日 | 2020-06-19 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Structural basis of GABARAP-mediated GABA A receptor trafficking and functions on GABAergic synaptic transmission.
Nat Commun, 12, 2021
|
|
7VI6
 
 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila | | 分子名称: | Beta-N-acetylhexosaminidase, CHLORIDE ION, MAGNESIUM ION | | 著者 | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | 登録日 | 2021-09-26 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7VI7
 
 | | Crystal structure of GH3 beta-N-acetylhexosaminidase Amuc_2109 from Akkermansia muciniphila in complex with GlcNAc | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | 著者 | Qian, K, Yang, W, Chen, X, Wang, Y, Zhang, M, Wang, M. | | 登録日 | 2021-09-26 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional and structural characterization of a GH3 beta-N-acetylhexosaminidase from Akkermansia muciniphila involved in mucin degradation
Biochem.Biophys.Res.Commun., 589, 2022
|
|
7V7M
 
 | | crystal structure of SARS-CoV-2 3CL protease | | 分子名称: | 3C-like proteinase | | 著者 | Yi, Y, Zhang, M, Ye, M. | | 登録日 | 2021-08-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Schaftoside inhibits 3CL pro and PL pro of SARS-CoV-2 virus and regulates immune response and inflammation of host cells for the treatment of COVID-19.
Acta Pharm Sin B, 12, 2022
|
|
7BQG
 
 | | Complex structure of SAV1 and Dendrin | | 分子名称: | POTASSIUM ION, Protein salvador homolog 1,Dendrin | | 著者 | Lin, Z, Zhang, M. | | 登録日 | 2020-03-24 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.55010867 Å) | | 主引用文献 | A WW Tandem-Mediated Dimerization Mode of SAV1 Essential for Hippo Signaling.
Cell Rep, 32, 2020
|
|
7FH5
 
 | | Structure of AdaV | | 分子名称: | AdaV, CHLORIDE ION, FE (III) ION | | 著者 | Zhang, Z.Y, Chen, W.Q, Zhai, G.Q, Zhang, M. | | 登録日 | 2021-07-29 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Insight into the Catalytic Mechanism of Non-Heme Iron Halogenase AdaV in 2'-Chloropentostatin Biosynthesis
Acs Catalysis, 12, 2022
|
|
7VYW
 
 | |
7V7W
 
 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex with oleoylethanolamide (OEA) | | 分子名称: | (Z)-N-(2-hydroxyethyl)octadec-9-enamide, Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | 著者 | Diao, X, Ren, X, Li, F.W, Zhang, M, Sun, X, Wu, D. | | 登録日 | 2021-08-21 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.507 Å) | | 主引用文献 | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
2FQD
 
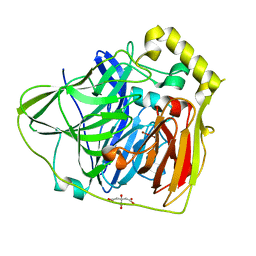 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | 分子名称: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | 著者 | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | 登録日 | 2006-01-18 | | 公開日 | 2007-01-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQE
 
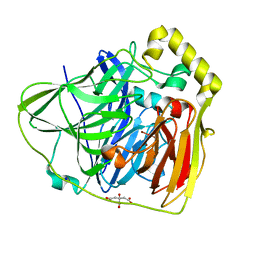 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | 分子名称: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | 著者 | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | 登録日 | 2006-01-18 | | 公開日 | 2007-01-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQG
 
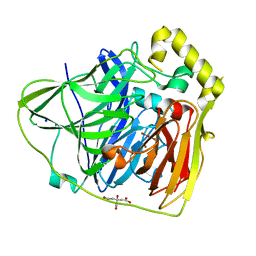 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | 分子名称: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | 著者 | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | 登録日 | 2006-01-18 | | 公開日 | 2007-01-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQF
 
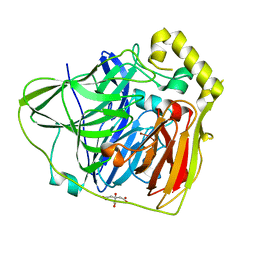 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | 分子名称: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | 著者 | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | 登録日 | 2006-01-18 | | 公開日 | 2007-01-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
3UT7
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | 分子名称: | Putative uncharacterized protein, SULFATE ION | | 著者 | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | 登録日 | 2011-11-25 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UT8
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | 分子名称: | IODIDE ION, Putative uncharacterized protein | | 著者 | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | 登録日 | 2011-11-25 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.168 Å) | | 主引用文献 | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
3UT4
 
 | | Structural view of a non Pfam singleton and crystal packing analysis | | 分子名称: | Putative uncharacterized protein | | 著者 | Cheng, C, Shaw, N, Zhang, X, Zhang, M, Ding, W, Wang, B.C, Liu, Z.J. | | 登録日 | 2011-11-25 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural view of a non pfam singleton and crystal packing analysis.
Plos One, 7, 2012
|
|
2HQ4
 
 | | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii | | 分子名称: | Hypothetical protein PH1570 | | 著者 | Li, Y, Marshall, M, Chang, J, Zhao, M, Zhang, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2006-07-18 | | 公開日 | 2006-09-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii
To be Published
|
|
4FYP
 
 | | Crystal Structure of Plant Vegetative Storage Protein | | 分子名称: | MAGNESIUM ION, Vegetative storage protein 1 | | 著者 | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | 登録日 | 2012-07-05 | | 公開日 | 2013-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
2IDG
 
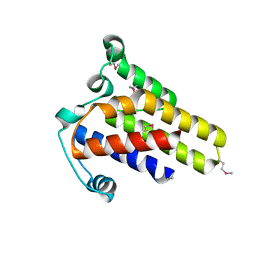 | | Crystal Structure of hypothetical protein AF0160 from Archaeoglobus fulgidus | | 分子名称: | Hypothetical protein AF0160 | | 著者 | Zhao, M, Zhang, M, Chang, J, Chen, L, Xu, H, Li, Y, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2006-09-15 | | 公開日 | 2006-11-14 | | 最終更新日 | 2017-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Crystal structure of Hypothetical Protein AF0160 from Archaeoglobus fulgidus at 2.69 Angstrom resolution
To be Published
|
|
2FJL
 
 | |
2FZ0
 
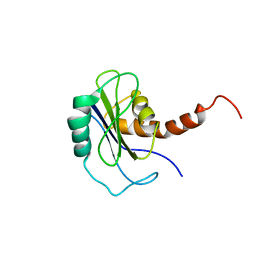 | | Identification of yeast R-SNARE Nyv1p as a novel longin domain protein | | 分子名称: | v-SNARE component of the vacuolar SNARE complex involved in vesicle fusion; inhibits ATP-dependent Ca(2+) transport activity of Pmc1p in the vacuolar membrane; Nyv1p | | 著者 | Wen, W, Zhang, M. | | 登録日 | 2006-02-08 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Identification of the Yeast R-SNARE Nyv1p as a Novel Longin Domain-containing Protein
Mol.Cell.Biol., 17, 2006
|
|
7F7I
 
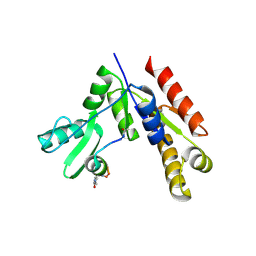 | | Stapled Peptide Inhibitor in complex with PSD95 GK domain | | 分子名称: | ACE-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYZ-ALA-ILE-GLN-NH2, Disks large homolog 4 | | 著者 | Shang, Y, Huang, X, Li, X, Zhang, M. | | 登録日 | 2021-06-29 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.595 Å) | | 主引用文献 | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
4D8O
 
 | |
4DC2
 
 | | Structure of PKC in Complex with a Substrate Peptide from Par-3 | | 分子名称: | ADENINE, Partitioning defective 3 homolog, Protein kinase C iota type | | 著者 | Shang, Y, Wang, C, Yu, J, Zhang, M. | | 登録日 | 2012-01-17 | | 公開日 | 2012-07-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Substrate recognition mechanism of atypical protein kinase Cs revealed by the structure of PKC iota in complex with a substrate peptide from Par-3
Structure, 20, 2012
|
|
4EDJ
 
 | | Crystal structure of the GRASP55 GRASP Domain with a phosphomimetic mutation (S189D) | | 分子名称: | Golgi reassembly-stacking protein 2, POTASSIUM ION | | 著者 | Truschel, S.T, Zhang, M, Bachert, C, Macbeth, M.R, Linstedt, A.D. | | 登録日 | 2012-03-27 | | 公開日 | 2012-05-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Allosteric Regulation of GRASP Protein-dependent Golgi Membrane Tethering by Mitotic Phosphorylation.
J.Biol.Chem., 287, 2012
|
|
