7S2Y
 
 | | SAMHD1 HD domain bound to CNDAC | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 4-amino-1-{2-cyano-2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}pyrimidin-2(1H)-one, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | 著者 | Digianantonio, K.M, Xiong, Y. | | 登録日 | 2021-09-04 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Differences between intrinsic and acquired nucleoside analogue resistance in acute myeloid leukaemia cells.
J Exp Clin Cancer Res, 40, 2021
|
|
6E49
 
 | | Pif1 peptide bound to PCNA trimer | | 分子名称: | ATP-dependent DNA helicase PIF1, Proliferating cell nuclear antigen | | 著者 | Buzovetsky, O, Kwon, Y, Pham, N.T, Kim, C, Ira, G, Sung, P, Xiong, Y. | | 登録日 | 2018-07-17 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Role of the Pif1-PCNA Complex in Pol delta-Dependent Strand Displacement DNA Synthesis and Break-Induced Replication.
Cell Rep, 21, 2017
|
|
6ECN
 
 | | HIV-1 CA 1/2-hexamer-EE | | 分子名称: | HIV-1 CA | | 著者 | Summers, B.J, Xiong, Y. | | 登録日 | 2018-08-08 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6EC2
 
 | | Structure of HIV-1 CA 1/3-hexamer | | 分子名称: | ACETATE ION, Capsid protein p24 | | 著者 | Summers, B.J, Xiong, Y. | | 登録日 | 2018-08-07 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6ECO
 
 | |
3DCG
 
 | | Crystal Structure of the HIV Vif BC-box in Complex with Human ElonginB and ElonginC | | 分子名称: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | 著者 | Stanley, B.J, Ehrlich, E.S, Short, L, Yu, Y, Xiao, Z, Yu, X.-F, Xiong, Y. | | 登録日 | 2008-06-03 | | 公開日 | 2008-07-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly
J.Virol., 82, 2008
|
|
4WQO
 
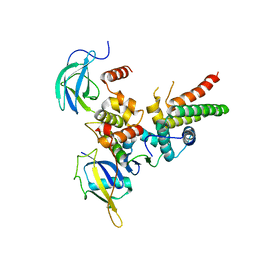 | | Structure of VHL-EloB-EloC-Cul2 | | 分子名称: | Cullin-2, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | 著者 | Nguyen, H.C, Xiong, Y. | | 登録日 | 2014-10-22 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Insights into Cullin-RING E3 Ubiquitin Ligase Recruitment: Structure of the VHL-EloBC-Cul2 Complex.
Structure, 23, 2015
|
|
8DZH
 
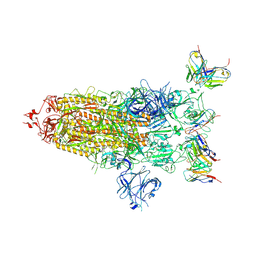 | |
8DZI
 
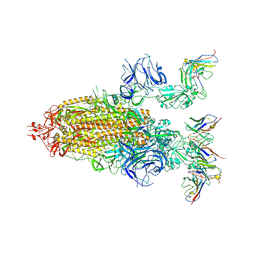 | |
8SZK
 
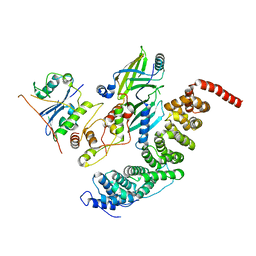 | |
8T4S
 
 | |
7N9U
 
 | | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions | | 分子名称: | Capsid protein, Nanobody | | 著者 | Gerber, E.E, Digianantonio, K.M, Tripler, T.N, Smaga, S.S, Summers, B.J, Xiong, Y. | | 登録日 | 2021-06-18 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
To Be Published
|
|
6PWY
 
 | | Structure of C. elegans ZK177.8, SAMHD1 ortholog | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Lim, C.S, Maehigashi, T, Wade, L.R, Bowen, N, Knecht, K, Xiong, Y, Kim, B. | | 登録日 | 2019-07-24 | | 公開日 | 2020-07-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | ZK177.8 of Caenorhabditis elegans:
Aicardi-Goutieres Syndrome SAMHD1 Ortholog
To Be Published
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | 分子名称: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | 著者 | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | 登録日 | 2016-09-29 | | 公開日 | 2016-12-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6QPL
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor MS31 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Johansson, C, Krojer, T, Xiong, Y, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | 登録日 | 2019-02-14 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery of a Potent and Selective Fragment-like Inhibitor of Methyllysine Reader Protein Spindlin 1 (SPIN1).
J.Med.Chem., 62, 2019
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | 分子名称: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | 著者 | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | 登録日 | 2016-09-29 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.755 Å) | | 主引用文献 | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
7UPN
 
 | |
7N9X
 
 | | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions | | 分子名称: | Capsid protein, Nanobody, Peptidyl-prolyl cis-trans isomerase A | | 著者 | Gerber, E.E, Digianantonio, K.M, Tripler, T.N, Smaga, S.S, Summers, B.J, Xiong, Y. | | 登録日 | 2021-06-18 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.511 Å) | | 主引用文献 | CA-targeting nanobody is a tool for studying HIV-1 capsid lattice interactions
To Be Published
|
|
7OCB
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor XY49-92B | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[3-(1,3-dihydroisoindol-2-yl)propoxy]-2N-[2-(dimethylamino)ethyl]-6-methoxy-4N-(1-propan-2-ylpiperidin-4-yl)quinazoline-2,4-diamine, CHLORIDE ION, ... | | 著者 | Johansson, C, Krojer, T, Park, K, Xiong, Y, Jin, J, Oppermann, U. | | 登録日 | 2021-04-26 | | 公開日 | 2022-05-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Crystal structure of Spindlin1 in complex with the inhibitor XY49-92B
To Be Published
|
|
1D9K
 
 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | 著者 | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | 登録日 | 1999-10-28 | | 公開日 | 1999-12-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
1EFO
 
 | | CRYSTAL STRUCTURE OF AN ADENINE BULGE IN THE RNA CHAIN OF A DNA/RNA HYBRID, D(CTCCTCTTC)/R(GAAGAGAGAG) | | 分子名称: | DNA (5'-D(*CP*TP*CP*CP*TP*CP*TP*TP*C)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3') | | 著者 | Sudarsanakumar, C, Xiong, Y, Sundaralingam, M. | | 登録日 | 2000-02-09 | | 公開日 | 2000-05-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of an adenine bulge in the RNA chain of a DNA.RNA hybrid, d(CTCCTCTTC).r(gaagagagag).
J.Mol.Biol., 299, 2000
|
|
3NCY
 
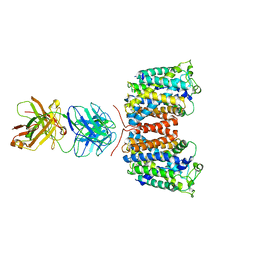 | | X-ray crystal structure of an arginine agmatine antiporter (AdiC) in complex with a Fab fragment | | 分子名称: | AdiC, Fab Heavy chain, Fab Light chain | | 著者 | Fang, Y, Jayaram, H, Shane, T, Komalkova-Partensky, L, Wu, F, Williams, C, Xiong, Y, Miller, C. | | 登録日 | 2010-06-06 | | 公開日 | 2010-08-18 | | 最終更新日 | 2019-07-17 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of a prokaryotic virtual proton pump at 3.2 A resolution.
Nature, 460, 2009
|
|
8WW2
 
 | | GPR3/Gs complex | | 分子名称: | CHOLESTEROL HEMISUCCINATE, G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | He, Y, Xiong, Y. | | 登録日 | 2023-10-24 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Identification of oleic acid as an endogenous ligand of GPR3.
Cell Res., 34, 2024
|
|
4ZWG
 
 | | Crystal structure of the GTP-dATP-bound catalytic core of SAMHD1 phosphomimetic T592E mutant | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Tang, C, Ji, X, Xiong, Y. | | 登録日 | 2015-05-19 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Impaired dNTPase Activity of SAMHD1 by Phosphomimetic Mutation of Thr-592.
J.Biol.Chem., 290, 2015
|
|
4ZWE
 
 | |
