8C6B
 
 | | Light SFX structure of D.m(6-4)photolyase at 20ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6F
 
 | | Light SFX structure of D.m(6-4)photolyase at 400fs time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
6WP8
 
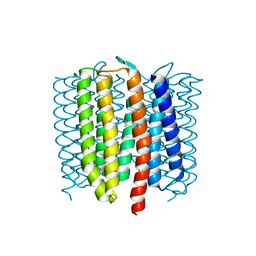 | | Proton-pumping mutant of Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | Proton-pumping rhodopsin chloride pump, RETINAL, octyl beta-D-glucopyranoside | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
6WZZ
 
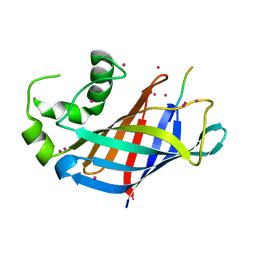 | | GID4 in complex with VGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION, VGLWKS peptide | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WZX
 
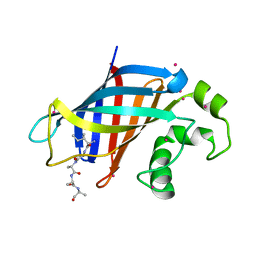 | | GID4 in complex with IGLWKS peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, ILE-GLY-LEU-TRP-LYS peptide, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-14 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | Authors: | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | Deposit date: | 2020-04-28 | | Release date: | 2021-06-09 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
6WIB
 
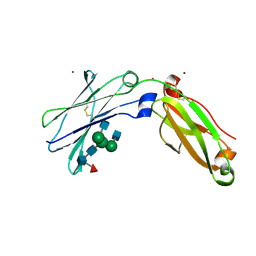 | | Next generation monomeric IgG4 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant gamma 4, ZINC ION | | Authors: | Oganesyan, V.Y, Shan, L, Dall'Acqua, W, van Dyk, N. | | Deposit date: | 2020-04-09 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6XL3
 
 | | Mastigocladopsis repens rhodopsin chloride pump | | Descriptor: | CHLORIDE ION, DECANE, Mastigocladopsis repens rhodopsin chloride pump, ... | | Authors: | Besaw, J.E, Ernst, O.P, Ou, W, Morizumi, T. | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The crystal structures of a chloride-pumping microbial rhodopsin and its proton-pumping mutant illuminate proton transfer determinants.
J.Biol.Chem., 295, 2020
|
|
6XRE
 
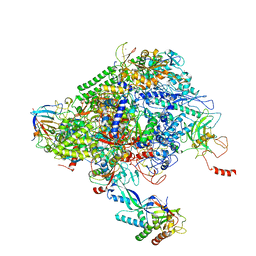 | | Structure of the p53/RNA polymerase II assembly | | Descriptor: | Cellular tumor antigen p53, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Liou, S.-H, Singh, S, Singer, R.H, Coleman, R.A, Liu, W. | | Deposit date: | 2020-07-12 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the p53/RNA polymerase II assembly.
Commun Biol, 4, 2021
|
|
6XOS
 
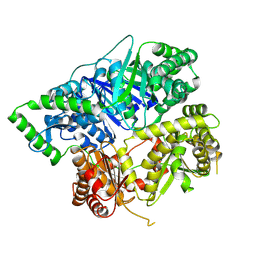 | |
6XOU
 
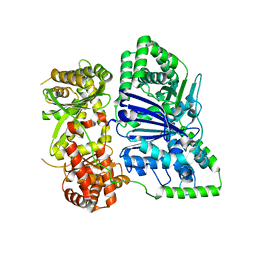 | |
6XOV
 
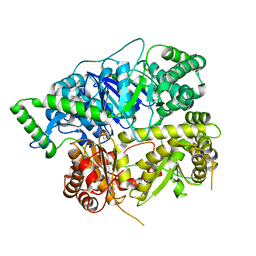 | |
6XOT
 
 | |
6XLY
 
 | |
6Y6D
 
 | | Tubulin-7-Aminonoscapine complex | | Descriptor: | (3~{S})-7-azanyl-6-methoxy-3-[(5~{R})-4-methoxy-6-methyl-7,8-dihydro-5~{H}-[1,3]dioxolo[4,5-g]isoquinolin-5-yl]-3~{H}-2-benzofuran-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Oliva, M.A, Prota, A.E, Rodriguez-Salarichs, J, Gu, W, Bennani, Y.L, Jimenez-Barbero, J, Canales, A, Steinmetz, M.O, Diaz, J.F. | | Deposit date: | 2020-02-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Noscapine Activation for Tubulin Binding.
J.Med.Chem., 63, 2020
|
|
4FOG
 
 | | Crystal Structure of Mtb ThyA in Complex with 5-Fluoro-dUMP and 5-methyltetrahydrofolic acid | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Thymidylate synthase | | Authors: | Reddy, M.C.M, Bruning, J.B, Harshbarger, W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-06-20 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of binary and ternary complexes of thymidylate synthase (ThyA) from Mycobacterium tuberculosis: Insights into the selectivity and mode of inhibition
To be Published
|
|
5QT8
 
 | | T. brucei FPPS in complex with CAS 34105-16-3 | | Descriptor: | (2R)-1-[(4-chlorophenyl)(methyl)amino]-3-(1H-imidazol-1-yl)propan-2-ol, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Jahnke, W. | | Deposit date: | 2019-08-07 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | T. brucei FPPS in complex with CAS 34105-16-3
To Be Published
|
|
6ZGX
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 1-[2-(2-oxidanylidenepyrrolidin-1-yl)ethyl]-3-phenyl-urea, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6ZGY
 
 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | (2,5-dimethylphenyl) pyridine-4-carboxylate, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6ZGW
 
 | | Structure of human galactokinase 1 bound with (4-chlorophenyl)methyl pyridine-3-carboxylate | | Descriptor: | (4-chlorophenyl)methyl pyridine-3-carboxylate, 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
5QTC
 
 | |
5QTD
 
 | | Unliganded T. brucei FPPS | | Descriptor: | Farnesyl pyrophosphate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Cornaciu, I, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-09 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Unliganded T. brucei FPPS
To Be Published
|
|
5QT5
 
 | | T. brucei FPPS in complex with CID 89021 | | Descriptor: | 6-ethylpyridin-2-amine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Jahnke, W. | | Deposit date: | 2019-08-07 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | T. brucei FPPS in complex with CID 89021
To Be Published
|
|
5QTE
 
 | | T. brucei FPPS in complex with CID 4563894 | | Descriptor: | (4S)-3-(4-chlorobenzene-1-carbonyl)-1,3-thiazolidine-4-carboxylic acid, Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Cornaciu, I, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-09 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | T. brucei FPPS in complex with CID 4563894
To Be Published
|
|
5QT4
 
 | | T. brucei FPPS in complex with CID 3599333 | | Descriptor: | 1-[3-(4-chlorophenoxy)propyl]-1H-imidazole, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Jahnke, W. | | Deposit date: | 2019-08-07 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | T. brucei FPPS in complex with CID 3599333
To Be Published
|
|
