191L
 
 | |
2E75
 
 | | Crystal Structure of the Cytochrome b6f Complex with 2-nonyl-4-hydroxyquinoline N-oxide (NQNO) from M.laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2-NONYL-4-HYDROXYQUINOLINE N-OXIDE, ... | | Authors: | Cramer, W.A, Yamashita, E, Zhang, H. | | Deposit date: | 2007-01-05 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of the Cytochrome b(6)f Complex: Quinone Analogue Inhibitors as Ligands of Heme c(n)
J.Mol.Biol., 370, 2007
|
|
6B0H
 
 | | Crystal structure of Pfs25 in complex with the transmission blocking antibody 1262 | | Descriptor: | 1,2-ETHANEDIOL, 1262 antibody, heavy chain, ... | | Authors: | McLeod, B, Scally, S.W, Bosch, A, King, C.R, Julien, J.P. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular definition of multiple sites of antibody inhibition of malaria transmission-blocking vaccine antigen Pfs25.
Nat Commun, 8, 2017
|
|
2YM8
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | (R)-5-(8-CHLOROISOQUINOLIN-3-YLAMINO)-3-(1-(DIMETHYLAMINO)PROPAN-2-YLOXY)PYRAZINE-2-CARBONITRILE, 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
1KZU
 
 | | INTEGRAL MEMBRANE PERIPHERAL LIGHT HARVESTING COMPLEX FROM RHODOPSEUDOMONAS ACIDOPHILA STRAIN 10050 | | Descriptor: | BACTERIOCHLOROPHYLL A, LIGHT HARVESTING PROTEIN B-800/850, Rhodopin b-D-glucoside | | Authors: | Cogdell, R.J, Freer, A.A, Isaacs, N.W, Hawthornthwaite-Lawless, A.M, Mcdermott, G, Papiz, M.Z, Prince, S.M. | | Deposit date: | 1996-08-31 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Apoprotein structure in the LH2 complex from Rhodopseudomonas acidophila strain 10050: modular assembly and protein pigment interactions.
J.Mol.Biol., 268, 1997
|
|
1S9F
 
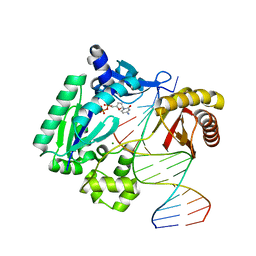 | | DPO with AT matched | | Descriptor: | 2',3'-DIDEOXYCYTOSINE-5'-DIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', 5'-D(*T*TP*CP*AP*GP*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Trincao, J, Johnson, R.E, Wolfle, W.T, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2004-02-04 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Nat.Struct.Mol.Biol., 11, 2004
|
|
2YKY
 
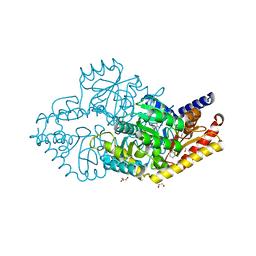 | | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase | | Descriptor: | (3S)-3-amino-3-phenylpropanoic acid, 1,2-ETHANEDIOL, BENZENE, ... | | Authors: | Wybenga, G.G, Crismaru, C.G, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2011-05-30 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Determinants of the Beta-Selectivity of a Bacterial Aminotransferase.
J.Biol.Chem., 287, 2012
|
|
6O9P
 
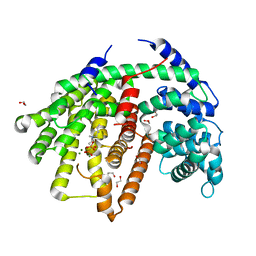 | | Wild-type SaSQS1 Complexed with Ibandronate | | Descriptor: | 1,2-ETHANEDIOL, IBANDRONATE, MAGNESIUM ION, ... | | Authors: | Blank, P.N, Christianson, D.W. | | Deposit date: | 2019-03-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Sesquisabinene Synthase 1, a Terpenoid Cyclase That Generates a Strained [3.1.0] Bridged-Bicyclic Product.
Acs Chem.Biol., 14, 2019
|
|
2YB4
 
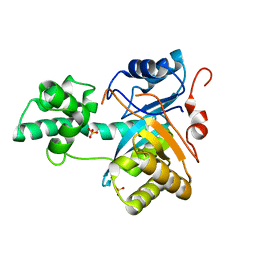 | | Structure of an amidohydrolase from Chromobacterium violaceum (EFI target EFI-500202) with bound SO4, no metal | | Descriptor: | AMIDOHYDROLASE, SULFATE ION | | Authors: | Vetting, M.W, Hillerich, B, Foti, R, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prospecting for Unannotated Enzymes: Discovery of a 3',5'-Nucleotide Bisphosphate Phosphatase within the Amidohydrolase Superfamily.
Biochemistry, 53, 2014
|
|
6AY4
 
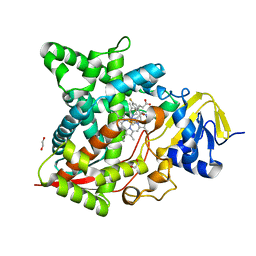 | | Naegleria fowleri CYP51-fluconazole complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, CYP51, ... | | Authors: | Debnath, A, Calvet, C.M, Jennings, G, Zhou, W, Aksenov, A, Luth, M, Abagyan, R, Nes, W.D, McKerrow, J.H, Podust, L.M. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CYP51 is an essential drug target for the treatment of primary amoebic meningoencephalitis (PAM).
PLoS Negl Trop Dis, 11, 2017
|
|
4EGN
 
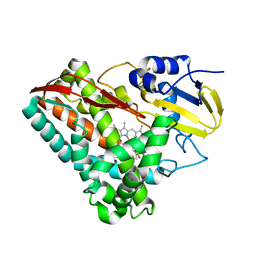 | | The X-ray crystal structure of CYP199A4 in complex with veratric acid | | Descriptor: | 3,4-dimethoxybenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
8DN5
 
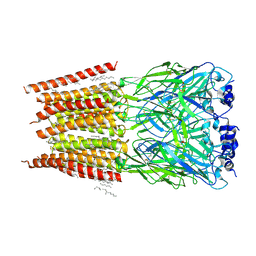 | |
8DN2
 
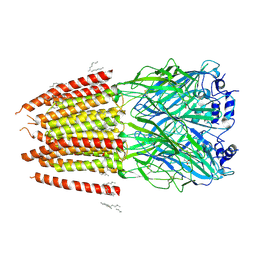 | |
8DN3
 
 | |
8DN4
 
 | | Cryo-EM structure of human Glycine Receptor alpha-1 beta heteromer, glycine-bound state3(desensitized state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-1, Glycine receptor subunit beta,Green fluorescent protein,Glycine receptor beta, ... | | Authors: | Liu, X, Wang, W. | | Deposit date: | 2022-07-10 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric gating of a human hetero-pentameric glycine receptor.
Nat Commun, 14, 2023
|
|
4JR5
 
 | | Structure-based Identification of Ureas as Novel Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(piperidin-1-ylsulfonyl)phenyl]-3-(pyridin-3-ylmethyl)thiourea, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Gunzner-Toste, J, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
3ZNN
 
 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 4H-THIENO[3,2-B]PYROLE-5-CARBOXYLIC ACID, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
1C69
 
 | |
1C6B
 
 | |
1C63
 
 | |
5KLV
 
 | | Structure of bos taurus cytochrome bc1 with fenamidone inhibited | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Zhou, Y, Xiao, Y, Tang, W.K, Yu, C.A, Qin, Z. | | Deposit date: | 2016-06-25 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
1C6C
 
 | |
7RBY
 
 | | Crystal structure of Nanobody nb112 and SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ilama-isolated nanobody NIH-CoV nb-112 specific to SARS-CoV-2 RBD, MAGNESIUM ION, ... | | Authors: | Chen, Y, Tolbert, W, Pazgier, M. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Nebulized delivery of a broadly neutralizing SARS-CoV-2 RBD-specific nanobody prevents clinical, virological, and pathological disease in a Syrian hamster model of COVID-19.
Mabs, 14
|
|
1C6D
 
 | |
3ZNP
 
 | | IN VITRO AND IN VIVO INHIBITION OF HUMAN D-AMINO ACID OXIDASE: REGULATION OF D-SERINE CONCENTRATION IN THE BRAIN | | Descriptor: | 3-HYDROXY-2H-CHROMEN-2-ONE, D-AMINO-ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hopkins, S.C, Heffernan, M.L.R, Saraswat, L.D, Bowen, C.A, Melnick, L, Hardy, L.W, Orsini, M.A, Allen, M.S, Koch, P, Spear, K.L, Foglesong, R.J, Soukri, M, Chytil, M, Fang, Q.K, Jones, S.W, Varney, M.A, Panatier, A, Oliet, S.H.R, Pollegioni, L, Piubelli, L, Molla, G, Nardini, M, Large, T.H. | | Deposit date: | 2013-02-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, Kinetic, and Pharmacodynamic Mechanisms of D-Amino Acid Oxidase Inhibition by Small Molecules.
J.Med.Chem., 56, 2013
|
|
