7TD4
 
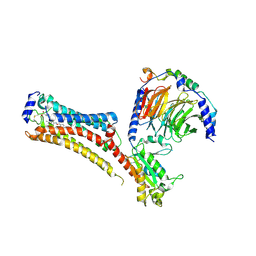 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to Siponimod | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7SNB
 
 | | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with AMP and ADP to 1.105 Angstrom resolution | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Fatty Acid Kinase A, ... | | Authors: | Cuypers, M.G, Subramanian, C, Rock, C.O, White, S.W. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Staphylococcus aureus Fatty Acid Kinase A (FakA, residues 1-208) in complex with AMP and ADP to 1.105 Angstrom resolution
To Be Published
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7SO6
 
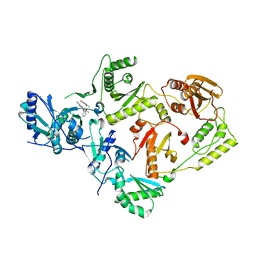 | | Crystal Structure of HIV-1 K103N, Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Bertoletti, N, Frey, K.M, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Chan, A.H. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7SO4
 
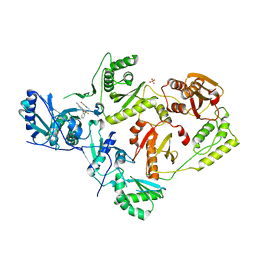 | | Crystal Structure of HIV-1 Y181C mutant Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Bertoletti, N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L, Frey, K.M, Chan, A.H. | | Deposit date: | 2021-10-29 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
7TEV
 
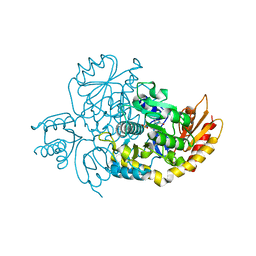 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (3S,4R)-3-amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylate | | Descriptor: | (1S,3R,4S)-3-formyl-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Zhu, W, Silverman, R, Liu, D. | | Deposit date: | 2022-01-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rational Design, Synthesis, and Mechanism of (3 S ,4 R )-3-Amino-4-(difluoromethyl)cyclopent-1-ene-1-carboxylic Acid: Employing a Second-Deprotonation Strategy for Selectivity of Human Ornithine Aminotransferase over GABA Aminotransferase.
J.Am.Chem.Soc., 144, 2022
|
|
7TDO
 
 | | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | Authors: | Liu, W, Schoonen, M, Wang, T, McSweeney, S, Liu, Q. | | Deposit date: | 2022-01-02 | | Release date: | 2022-04-06 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of transmembrane AAA+ protease FtsH in the ADP state.
Commun Biol, 5, 2022
|
|
7SWO
 
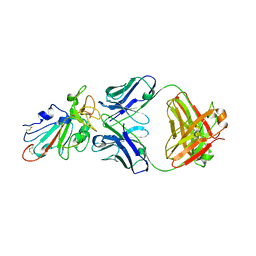 | | C98C7 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | C98C7 Fab heavy chain, C98C7 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWP
 
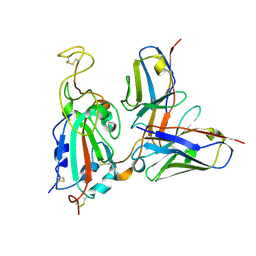 | | G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32Q4 Fab heavy chain, G32Q4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWN
 
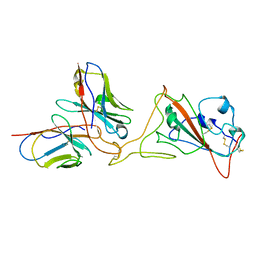 | | G32A4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32A4 Fab heavy chain, G32A4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7T1T
 
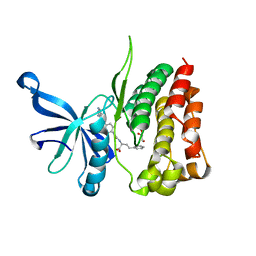 | | JAK2 JH2 IN COMPLEX WITH JAK292 | | Descriptor: | (2S)-2-[({4-[(2-amino-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}carbamoyl)amino]-4-phenylbutanoic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Ippolito, J.A, Henry, S, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2021-12-02 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Conversion of a False Virtual Screen Hit into Selective JAK2 JH2 Domain Binders Using Convergent Design Strategies
Acs Med.Chem.Lett., 13, 2022
|
|
7SQY
 
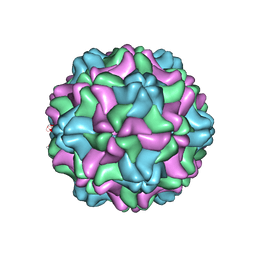 | | CSDaV GFP mutant | | Descriptor: | Citrus Sudden Death-associated Virus Capsid Protein,Green fluorescent protein,Citrus Sudden Death-associated Virus Capsid Protein | | Authors: | Guo, F, Matsumura, E.E, Falk, B.W. | | Deposit date: | 2021-11-07 | | Release date: | 2022-05-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Citrus sudden death-associated virus as a new expression vector for rapid in planta production of heterologous proteins, chimeric virions, and virus-like particles.
Biotechnol Rep., 35, 2022
|
|
5XD0
 
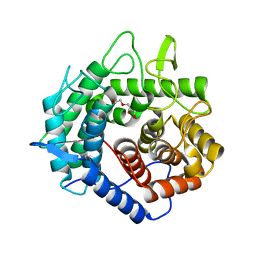 | | Apo Structure of Beta-1,3-1,4-glucanase from Paenibacillus sp.X4 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glucanase, TRIETHYLENE GLYCOL | | Authors: | Baek, S.C, Ho, T.-H, Kang, L.-W, Kim, H. | | Deposit date: | 2017-03-24 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Improvement of enzyme activity of beta-1,3-1,4-glucanase from Paenibacillus sp. X4 by error-prone PCR and structural insights of mutated residues.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
7STQ
 
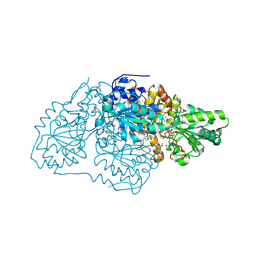 | | Crystal structure of arabidopsis thaliana acetohydroxyacid synthase W574L mutant in complex with chlorimuron-ethyl | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2021-11-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of resistance to herbicides that target acetohydroxyacid synthase.
Nat Commun, 13, 2022
|
|
7T0P
 
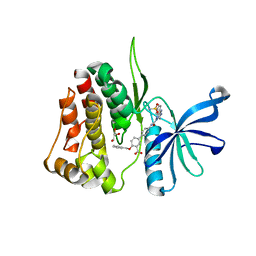 | | JAK2 JH2 IN COMPLEX WITH JAK315 | | Descriptor: | 4'-{[5-amino-3-(4-sulfamoylanilino)-1H-1,2,4-triazole-1-carbonyl]amino}-4-(benzyloxy)[1,1'-biphenyl]-3-carboxylic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Ippolito, J.A, Liosi, M.-E, Krimmer, S.G, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2021-11-30 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights on JAK2 Modulation by Potent, Selective, and Cell-Permeable Pseudokinase-Domain Ligands.
J.Med.Chem., 65, 2022
|
|
7SU9
 
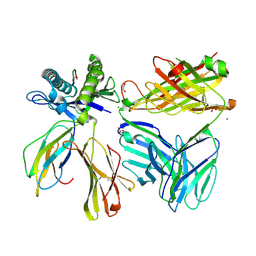 | |
7SW4
 
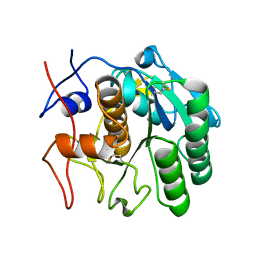 | | MicroED structure of proteinase K from a 540 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.4 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW9
 
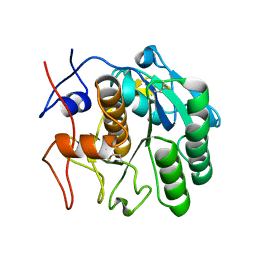 | | MicroED structure of proteinase K from a 170 nm thick lamella measured at 300 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SVY
 
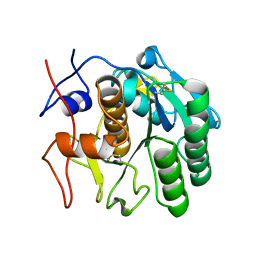 | | MicroED structure of proteinase K from a 130 nm thick lamella measured at 120 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.3 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SWB
 
 | | MicroED structure of proteinase K from a 360 nm thick lamella measured at 300 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.05 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW2
 
 | | MicroED structure of proteinase K from a 130 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.95 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW8
 
 | | MicroED structure of proteinase K from a 150 nm thick lamella measured at 300 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW5
 
 | | MicroED structure of proteinase K from a 460 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.95 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SVZ
 
 | | MicroED structure of proteinase K from a 200 nm thick lamella measured at 120 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
