8UAT
 
 | | Thermus scotoductus SA-01 Ene-reductase Compound 3b Complex | | Descriptor: | 1-[2-(4-hydroxyphenyl)ethyl]-1,4-dihydropyridine-3-carboxamide, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wilson, L.A, Guddat, L.W, Schenk, G, Scott, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
2JJ2
 
 | | The Structure of F1-ATPase inhibited by quercetin. | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
7UWD
 
 | | Citrus V-ATPase State 2, H in contact with subunits AB | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit, V-type proton ATPase subunit AP1 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
7UTI
 
 | | ALTERNATIVE MODELING OF TROPOMYOSIN IN HUMAN CARDIAC THIN FILAMENT IN THE CALCIUM BOUND STATE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rynkiewicz, M.J, Pavadai, E, Lehman, W. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Protein-Protein Docking Reveals Dynamic Interactions of Tropomyosin on Actin Filaments.
Biophys J, 119, 2020
|
|
1LCP
 
 | |
4EHG
 
 | | B-Raf Kinase Domain in Complex with an Aminopyridimine-based Inhibitor | | Descriptor: | N-{2,4-difluoro-3-[({6-[(2-hydroxyethyl)amino]pyrimidin-4-yl}carbamoyl)amino]phenyl}propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Voegtli, W.C. | | Deposit date: | 2012-04-02 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent and selective aminopyrimidine-based B-raf inhibitors with favorable physicochemical and pharmacokinetic properties.
J.Med.Chem., 55, 2012
|
|
7UWC
 
 | | Citrus V-ATPase State 2, H in contact with subunit a | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit AP1 fragment, V-type proton ATPase subunit AP2 fragment, ... | | Authors: | Keon, K.A, Abdelaziz, R.A, Schulze, W.X, Schumacher, K, Rubinstein, J.L. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of V-ATPase from citrus fruit.
Structure, 30, 2022
|
|
6E1X
 
 | | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-10 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To Be Published
|
|
4LB1
 
 | |
1FTW
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | (5S,7R,8S,9S,10R)-3,8,9,10-tetrahydroxy-7-(hydroxymethyl)-6-oxa-1,3-diazaspiro[4.5]decane-2,4-dione, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
6Z17
 
 | | PqsR (MvfR) in complex with antagonist 6 | | Descriptor: | 6-chloranyl-3-[(2-propan-2-yl-2,3-dihydro-1,3-thiazol-4-yl)methyl]quinazolin-4-one, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2020-05-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Novel quinazolinone inhibitors of the Pseudomonas aeruginosa quorum sensing transcriptional regulator PqsR.
Eur.J.Med.Chem., 208, 2020
|
|
6JZL
 
 | |
3T7B
 
 | | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis | | Descriptor: | Acetylglutamate kinase, GLUTAMIC ACID, S,R MESO-TARTARIC ACID | | Authors: | Demas, M.W, Solberg, R.G, Cooper, D.R, Chruszcz, M, Porebski, P.J, Zheng, H, Onopriyenko, O, Skarina, T, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-29 | | Release date: | 2011-09-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis
To be Published
|
|
4LA6
 
 | |
1L0H
 
 | | CRYSTAL STRUCTURE OF BUTYRYL-ACP FROM E.COLI | | Descriptor: | ACYL CARRIER PROTEIN, ZINC ION | | Authors: | Roujeinikova, A, Baldock, C, Simon, W.J, Gilroy, J, Baker, P.J, Stuitje, A.R, Rice, D.W, Slabas, A.R, Rafferty, J.B. | | Deposit date: | 2002-02-11 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site
Structure, 10, 2002
|
|
1FTQ
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | (5S,7R,8S,9S,10R)-3-amino-8,9,10-trihydroxy-7-(hydroxymethyl)-6-oxa-1,3-diazaspiro[4.5]decane-2,4-dione, GLYCOGEN PHOSPHORYLASE, INOSINIC ACID, ... | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
5UNN
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in apo form | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH-dependent glyoxylate/hydroxypyruvate reductase | | Authors: | Shabalin, I.G, LaRowe, C, Kutner, J, Gasiorowska, O.A, Handing, K.B, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V0H
 
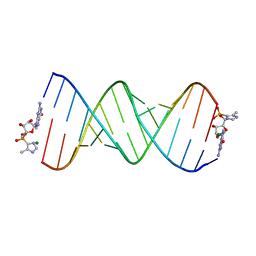 | | RNA duplex with 2-MeImpG analogue bound-one binding site | | Descriptor: | 5'-O-[(S)-hydroxy(4-methyl-1H-imidazol-5-yl)phosphoryl]guanosine, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*GP*G)-3') | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
J. Am. Chem. Soc., 140, 2018
|
|
1CZV
 
 | | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V: DIMERIC CRYSTAL FORM | | Descriptor: | PROTEIN (COAGULATION FACTOR V) | | Authors: | Macedo-Ribeiro, S, Bode, W, Huber, R, Kane, W.H, Fuentes-Prior, P. | | Deposit date: | 1999-09-07 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the membrane-binding C2 domain of human coagulation factor V.
Nature, 402, 1999
|
|
2JJ1
 
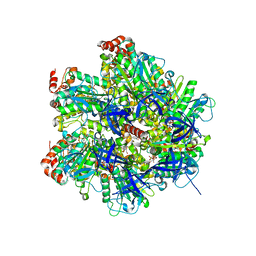 | | The Structure of F1-ATPase inhibited by piceatannol. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE GAMMA CHAIN, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, ... | | Authors: | Gledhill, J.R, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Inhibition of Bovine F1-ATPase by Resveratrol and Related Polyphenols.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6UID
 
 | |
1Y9D
 
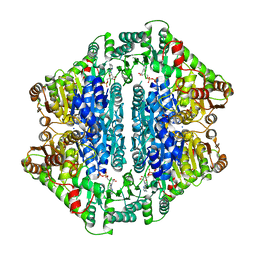 | | Pyruvate Oxidase variant V265A from Lactobacillus plantarum | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Pyruvate oxidase, ... | | Authors: | Wille, G, Ritter, M, Weiss, M.S, Konig, S, Mantele, W, Hubner, G. | | Deposit date: | 2004-12-15 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The role of Val-265 for Flavin Adenine Dinulceotide (FAD) binding in pyruvate oxidase: FTIR, kinetic and crystallographic studies on the enzyme variant V265A
Biochemistry, 44, 2005
|
|
6KKM
 
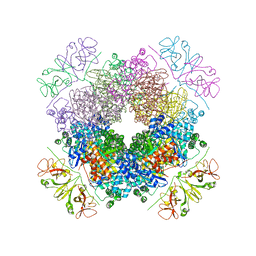 | | Crystal structure of RbcL-Raf1 complex from Anabaena sp. PCC 7120 | | Descriptor: | All5250 protein, Ribulose bisphosphate carboxylase large chain | | Authors: | Xia, L.Y, Jiang, Y.L, Kong, W.W, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-07-26 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for the assembly of RuBisCO assisted by the chaperone Raf1.
Nat.Plants, 6, 2020
|
|
5GM9
 
 | | Crystal structure of a glycoside hydrolase in complex with cellobiose | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-13 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
1G2V
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TTP COMPLEX. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
