5XJX
 
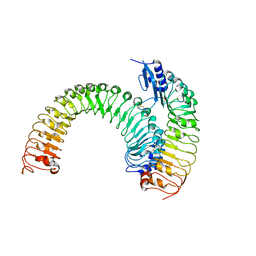 | | Pre-formed plant receptor ERL1-TMM complex | | Descriptor: | LRR receptor-like serine/threonine-protein kinase ERL1, Protein TOO MANY MOUTHS | | Authors: | Chai, J, Lin, G, Zhang, L, Han, Z, Yang, X, Liu, W, Qi, Y, Chang, J, Li, E. | | Deposit date: | 2017-05-04 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.055 Å) | | Cite: | A receptor-like protein acts as a specificity switch for the regulation of stomatal development.
Genes Dev., 31, 2017
|
|
8SB6
 
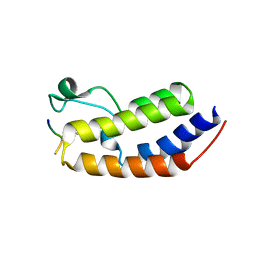 | | Structure of human BRD2-BD1 bound to a histone H4 acetyl-methyllysine peptide | | Descriptor: | Bromodomain containing 2, Histone H4 | | Authors: | Connor, L.J, Ekundayo, B.E, Lu-Culligan, W.J, Simon, M.D, Bleichert, F. | | Deposit date: | 2023-04-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acetyl-methyllysine marks chromatin at active transcription start sites.
Nature, 622, 2023
|
|
1YRQ
 
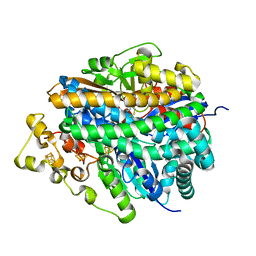 | | Structure of the ready oxidized form of [NiFe]-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Cavazza, C, Matho, M, Faber, B.W, Roseboom, W, Albracht, S.P, Garcin, E, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2005-02-04 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases.
J.Biol.Inorg.Chem., 10, 2005
|
|
2VY9
 
 | | Molecular architecture of the stressosome, a signal integration and transduction hub | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST | | Authors: | Marles-Wright, J, Grant, T, Delumeau, O, van Duinen, G, Firbank, S.J, Lewis, P.J, Murray, J.W, Newman, J.A, Quin, M.B, Race, P.R, Rohou, A, Tichelaar, W, van Heel, M, Lewis, R.J. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Architecture of the "Stressosome," a Signal Integration and Transduction Hub
Science, 322, 2008
|
|
8SCL
 
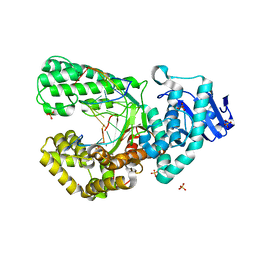 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 6h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8K6Z
 
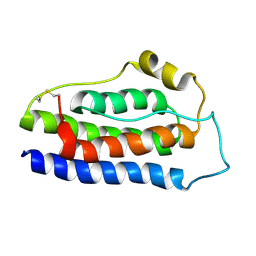 | | NMR structure of human leptin | | Descriptor: | Leptin | | Authors: | Fan, X, Qin, R, Yuan, W, Fan, J, Huang, W, Lin, Z. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Structure, 32, 2024
|
|
8SCN
 
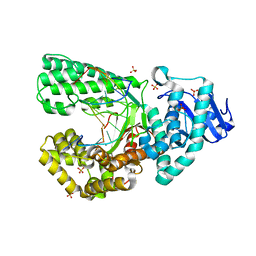 | | Bst DNA polymerase I Large Fragment mutant F710Y/D598A with 3'-amino primer, dGTP, and calcium time-resolved 24h (Product State) | | Descriptor: | CALCIUM ION, DIPHOSPHATE, DNA polymerase I, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCP
 
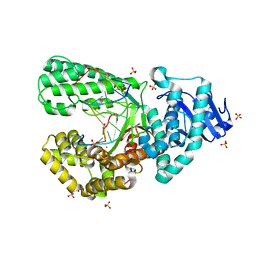 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 1h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
5XV1
 
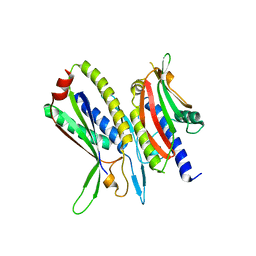 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Crystal structure of ATG101-ATG13HORMA
To Be Published
|
|
8SCQ
 
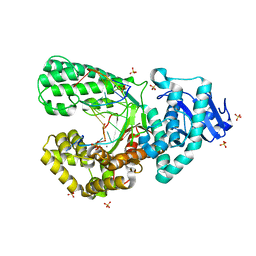 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 2h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCR
 
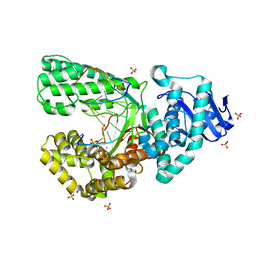 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 4h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCS
 
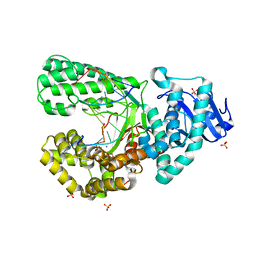 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 8h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCT
 
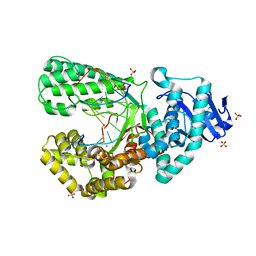 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 24h | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
8SCU
 
 | | Bst DNA polymerase I Large Fragment wildtype D598A with 3'-amino primer, dGTP, and calcium time-resolved 48h (Product State) | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DIPHOSPHATE, ... | | Authors: | Fang, Z, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Trivalent rare earth metal cofactors confer rapid NP-DNA polymerase activity.
Science, 382, 2023
|
|
6AK4
 
 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (~{E})-2-cyano-~{N},~{N}-diethyl-3-[3-nitro-4,5-bis(oxidanyl)phenyl]prop-2-enamide, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
4QDR
 
 | |
6UJH
 
 | |
2KF8
 
 | | Structure of a two-G-tetrad basket-type intramolecular G-quadruplex formed by human telomeric repeats in K+ solution | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Lim, K.W, Amrane, S, Bouaziz, S, Xu, W, Mu, Y, Patel, D.J, Luu, K.N, Phan, A.T. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers
J.Am.Chem.Soc., 131, 2009
|
|
2YC5
 
 | | Inhibitors of the herbicidal target IspD | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CHLOROPLASTIC, 5-chloro-7-hydroxy-6-(phenylmethyl)pyrazolo[1,5-a]pyrimidine-3-carbonitrile, ... | | Authors: | Hoeffken, H.W. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitors of the Herbicidal Target Ispd: Allosteric Site Binding.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
1LRA
 
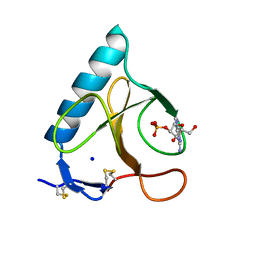 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
7UQA
 
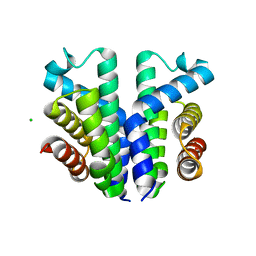 | | Crystal structure of the small Ultra-Red Fluorescent Protein (smURFP) | | Descriptor: | CHLORIDE ION, SODIUM ION, small Ultra-Red Fluorescent Protein (smURFP) | | Authors: | Maiti, A, Buffalo, C.Z, Saurabh, S, Montecinos-Franjola, F, Hachey, J.S, Conlon, W.J, Tran, G.N, Drobizhev, M, Moerner, W.E, Ghosh, P, Matsuo, H, Tsien, R.Y, Lin, J.Y, Rodriguez, E.A. | | Deposit date: | 2022-04-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structural and photophysical characterization of the small ultra-red fluorescent protein.
Nat Commun, 14, 2023
|
|
6B70
 
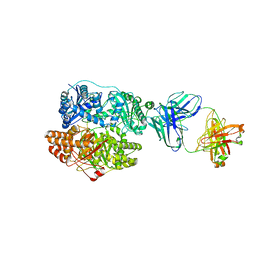 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin | | Descriptor: | FAB H11-E heavy chain, FAB H11-E light chain, Insulin, ... | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
8RHL
 
 | | Yeast 20S proteasome in complex with a linear biarylether epoxyketone (compound 15a) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Linear biarylether epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
8RHJ
 
 | | Yeast 20S proteasome in complex with a macrocyclic oxindole epoxyketone (compound 5) | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Macrocyclic oxindole epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
5XSE
 
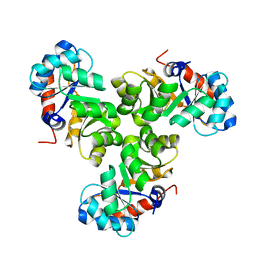 | |
