2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | 著者 | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | 登録日 | 2011-07-26 | | 公開日 | 2011-09-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
6TBI
 
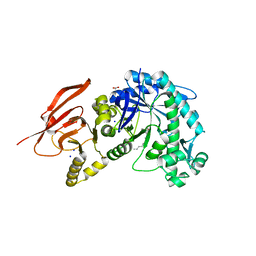 | | Structure of a beta galactosidase with inhibitor | | 分子名称: | (1S,2S,3S,6R)-4-(hydroxymethyl)-6-(octylamino)cyclohex-4-ene-1,2,3-triol, ACETATE ION, Beta-galactosidase, ... | | 著者 | Offen, W, Davies, G. | | 登録日 | 2019-11-01 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
5S4J
 
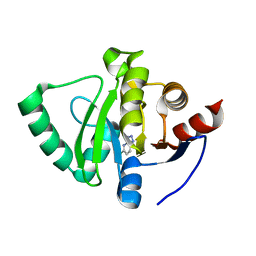 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF054 | | 分子名称: | 6-chlorotetrazolo[1,5-b]pyridazine, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.124 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2V85
 
 | |
3L29
 
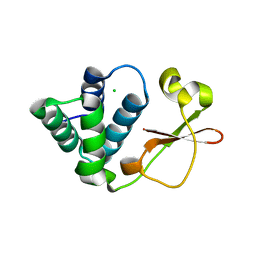 | | Crystal Structure of Zaire Ebola VP35 interferon inhibitory domain K319A/R322A mutant | | 分子名称: | CHLORIDE ION, Polymerase cofactor VP35 | | 著者 | Leung, D.W, Ramanan, P, Borek, D.M, Amarasinghe, G.K. | | 登録日 | 2009-12-14 | | 公開日 | 2010-02-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mutations abrogating VP35 interaction with double-stranded RNA render ebola virus avirulent in guinea pigs.
J.Virol., 84, 2010
|
|
7WPA
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer complexed with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | 登録日 | 2022-01-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WP9
 
 | | SARS-CoV-2 Omicron Variant SPIKE trimer, all RBDs down | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S, Xu, E. | | 登録日 | 2022-01-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
7WPD
 
 | | SARS-CoV-2 Omicron Variant S Trimer complexed with one JMB2002 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-Fab nanobody, ... | | 著者 | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | 登録日 | 2022-01-23 | | 公開日 | 2022-03-16 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
2V86
 
 | |
2HMG
 
 | | REFINEMENT OF THE INFLUENZA VIRUS HEMAGGLUTININ BY SIMULATED ANNEALING | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ (HA1 CHAIN), HEMAGGLUTININ (HA2 CHAIN), ... | | 著者 | Weis, W.I, Bruenger, A.T, Skehel, J.J, Wiley, D.C. | | 登録日 | 1989-09-11 | | 公開日 | 1991-01-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Refinement of the influenza virus hemagglutinin by simulated annealing.
J.Mol.Biol., 212, 1990
|
|
2HOA
 
 | | STRUCTURE DETERMINATION OF THE ANTP(C39->S) HOMEODOMAIN FROM NUCLEAR MAGNETIC RESONANCE DATA IN SOLUTION USING A NOVEL STRATEGY FOR THE STRUCTURE CALCULATION WITH THE PROGRAMS DIANA, CALIBA, HABAS AND GLOMSA | | 分子名称: | ANTENNAPEDIA PROTEIN | | 著者 | Guntert, P, Qian, Y.-Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | 登録日 | 1992-04-04 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure determination of the Antp (C39----S) homeodomain from nuclear magnetic resonance data in solution using a novel strategy for the structure calculation with the programs DIANA, CALIBA, HABAS and GLOMSA.
J.Mol.Biol., 217, 1991
|
|
224L
 
 | |
5S4I
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF051 | | 分子名称: | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.131 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4H
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF048 | | 分子名称: | 1-carbamoylpiperidine-4-carboxylic acid, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.175 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4F
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF003 | | 分子名称: | 1,8-naphthyridine, Non-structural protein 3, SULFATE ION | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.131 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
2XPV
 
 | | TetR(D) in complex with minocycline and magnesium. | | 分子名称: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Dalm, D, Proft, J, Palm, G.J, Hinrichs, W. | | 登録日 | 2010-08-30 | | 公開日 | 2011-09-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Tetr(D) in Complex with Minocycline.
To be Published
|
|
5S4G
 
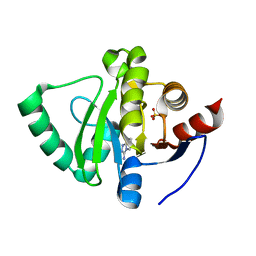 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF005 | | 分子名称: | Non-structural protein 3, SULFATE ION, [1,2,4]triazolo[4,3-a]pyridin-3-amine | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.172 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4R3O
 
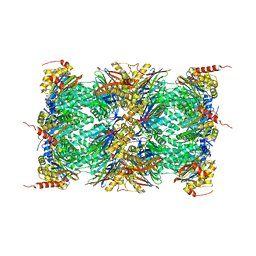 | | Human Constitutive 20S Proteasome | | 分子名称: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | 著者 | Sacchettini, J.C, Harshbarger, W.H. | | 登録日 | 2014-08-16 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of the Human 20S Proteasome in Complex with Carfilzomib.
Structure, 23, 2015
|
|
3BKC
 
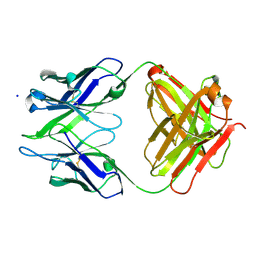 | | Crystal structure of anti-amyloid beta FAB WO2 (P21, FormB) | | 分子名称: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | 著者 | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | 登録日 | 2007-12-06 | | 公開日 | 2008-04-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | 分子名称: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | 著者 | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | 登録日 | 2021-06-17 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (37 Å) | | 主引用文献 | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
5FL2
 
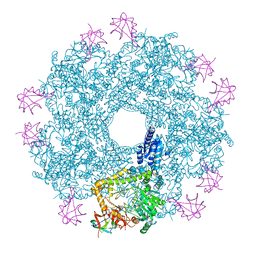 | | Revisited cryo-EM structure of Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | | 分子名称: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | 著者 | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | 登録日 | 2015-10-21 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
3IYM
 
 | | Backbone Trace of the Capsid Protein Dimer of a Fungal Partitivirus from Electron Cryomicroscopy and Homology Modeling | | 分子名称: | Capsid protein | | 著者 | Tang, J, Pan, J, Havens, W.F, Ochoa, W.F, Li, H, Sinkovits, R.S, Guu, T.S.Y, Ghabrial, S.A, Nibert, M.L, Tao, J.Y, Baker, T.S. | | 登録日 | 2010-02-05 | | 公開日 | 2010-07-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Backbone Trace of Partitivirus Capsid Protein from Electron Cryomicroscopy and Homology Modeling
Biophys.J., 99, 2010
|
|
5HQ8
 
 | |
3LJR
 
 | | GLUTATHIONE TRANSFERASE (THETA CLASS) FROM HUMAN IN COMPLEX WITH THE GLUTATHIONE CONJUGATE OF 1-MENAPHTHYL SULFATE | | 分子名称: | 1-MENAPHTHYL GLUTATHIONE CONJUGATE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | 著者 | Rossjohn, J, Mckinstry, W.J, Oakley, A.J, Verger, D, Flanagan, J, Chelvanayagam, G, Tan, K.L, Board, P.G, Parker, M.W. | | 登録日 | 1998-03-08 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site.
Structure, 6, 1998
|
|
3C7Q
 
 | | Structure of VEGFR2 kinase domain in complex with BIBF1120 | | 分子名称: | SULFATE ION, Vascular endothelial growth factor receptor 2, methyl (3Z)-3-{[(4-{methyl[(4-methylpiperazin-1-yl)acetyl]amino}phenyl)amino](phenyl)methylidene}-2-oxo-2,3-dihydro-1H-indole-6-carboxylate | | 著者 | Hilberg, F, Roth, G.J, Krssak, M, Kautschitsch, S, Sommergruber, W, Tontsch-Grunt, U, Garin-Chesa, P, Bader, G, Zoephel, A, Quant, J, Heckel, A, Rettig, W.J. | | 登録日 | 2008-02-08 | | 公開日 | 2008-12-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | BIBF 1120: triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy.
Cancer Res., 68, 2008
|
|
