2HIM
 
 | | Crystal Structure and Allosteric Regulation of the Cytoplasmic Escherichia coli L-Asparaginase I | | Descriptor: | 1,2-ETHANEDIOL, ASPARAGINE, ASPARTIC ACID, ... | | Authors: | Yun, M.K, Nourse, A, White, S.W, Rock, C.O, Heath, R.J. | | Deposit date: | 2006-06-29 | | Release date: | 2007-05-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure and Allosteric Regulation of the Cytoplasmic Escherichia coli L-Asparaginase I.
J.Mol.Biol., 369, 2007
|
|
6RRD
 
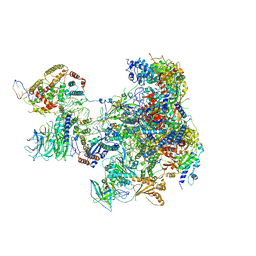 | | RNA Polymerase I Pre-initiation complex DNA opening intermediate 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-05-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
3S71
 
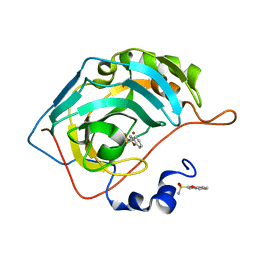 | |
2HR7
 
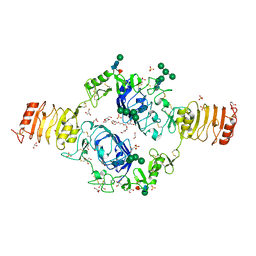 | | Insulin receptor (domains 1-3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garrett, T.P.J, Ward, C.W. | | Deposit date: | 2006-07-19 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The first three domains of the insulin receptor differ structurally from the insulin-like growth factor 1 receptor in the regions governing ligand specificity.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6DKC
 
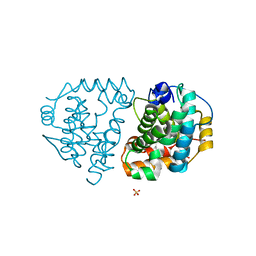 | | Yeast Ddi2 Cyanamide Hydratase, T157V mutant, apo structure | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
5T5G
 
 | | human SETD8 in complex with MS2177 | | Descriptor: | 7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)-N-[5-(pyrrolidin-1-yl)pentyl]quinazolin-4-amine, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Yu, W, Tempel, W, Babault, N, Ma, A, Butler, K.V, Jin, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of a Covalent Inhibitor of the SET Domain-Containing Protein 8 (SETD8) Lysine Methyltransferase.
J. Med. Chem., 59, 2016
|
|
2HJG
 
 | | The crystal structure of the B. subtilis YphC GTPase in complex with GDP | | Descriptor: | GTP-binding protein engA, GUANOSINE-5'-DIPHOSPHATE, ZINC ION | | Authors: | Muench, S.P, Xu, L, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The essential GTPase YphC displays a major domain rearrangement associated with nucleotide binding.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1FSP
 
 | | NMR SOLUTION STRUCTURE OF BACILLUS SUBTILIS SPO0F PROTEIN, 20 STRUCTURES | | Descriptor: | STAGE 0 SPORULATION PROTEIN F | | Authors: | Feher, V.A, Skelton, N.J, Dahlquist, F.W, Cavanagh, J. | | Deposit date: | 1997-06-05 | | Release date: | 1997-12-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Biochemistry, 36, 1997
|
|
6DOH
 
 | |
1RJP
 
 | | Crystal structure of D-aminoacylase in complex with 100mM CuCl2 | | Descriptor: | ACETATE ION, COPPER (II) ION, D-aminoacylase, ... | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
6DPF
 
 | |
5HIO
 
 | |
5T4I
 
 | |
5T62
 
 | | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3-Tif6-Lsg1 Complex | | Descriptor: | 25S Ribosomal RNA, 5.8S Ribosomal RNA, 5S Ribosomal RNA, ... | | Authors: | Malyutin, A.G, Musalgaonkar, S, Patchett, S, Frank, J, Johnson, A.W. | | Deposit date: | 2016-09-01 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis.
EMBO J., 36, 2017
|
|
7UE8
 
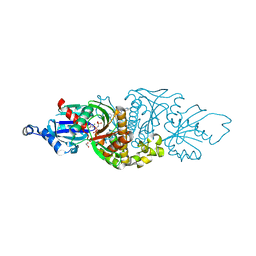 | | PANK3 complex structure with compound PZ-3890 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(6-chloropyridazin-3-yl)piperazin-1-yl]-2-(4-cyclopropyl-3-fluorophenyl)ethan-1-one, ACETATE ION, ... | | Authors: | White, S.W, Yun, M, Lee, R.E. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Relief of CoA sequestration and restoration of mitochondrial function in a mouse model of propionic acidemia.
J Inherit Metab Dis, 46, 2023
|
|
5T5C
 
 | |
5EHA
 
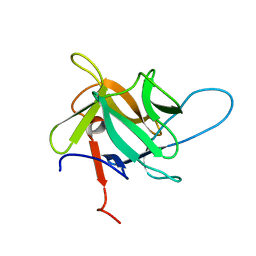 | | Crystal structure of recombinant MtaL at 1.35 Angstrom resolution | | Descriptor: | Lectin-like fold protein | | Authors: | Lai, X.-L, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of recombinant tyrosinase-binding protein MtaL at 1.35 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5ER8
 
 | |
7MOS
 
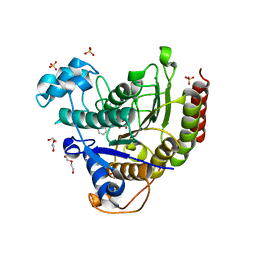 | | Structure of HDAC2 in complex with a macrocyclic inhibitor (compound 4) | | Descriptor: | (3S,18S,20aR)-18-(6,6-dihydroxyoctyl)-1,5,6,7,8,18,19,20a-octahydro-4H-14,17-epiminoazeto[1,2-g][1,7,10,13]benzoxatriazacycloheptadecin-20(2H)-one, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOX
 
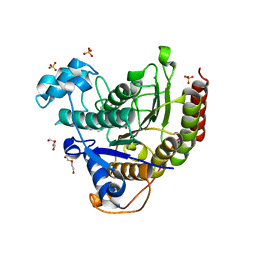 | | Structure of HDAC2 in complex with an inhibitor (compound 14) | | Descriptor: | (1S)-N-[(1S)-7,7-dihydroxy-1-{4-[(1R,4S)-1,2,3,4-tetrahydro-1,4-methanonaphthalen-6-yl]-1H-imidazol-2-yl}nonyl]-6-methyl-6-azaspiro[2.5]octane-1-carboxamide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7MOZ
 
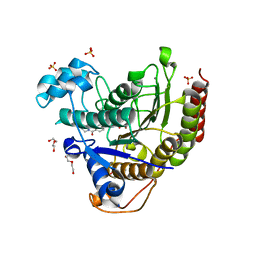 | | Structure of HDAC2 in complex with a macrocyclic inhibitor (compound 25) | | Descriptor: | (1R,3S,6S,18R,27R)-6-(6,6-dihydroxyoctyl)-5,8,18,27,34-pentaazahexacyclo[25.2.2.1~7,10~.1~11,15~.1~14,18~.0~1,3~]tetratriaconta-7,9,11(33),12,14,16-hexaene-4,32-dione (non-preferred name), CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
6RUI
 
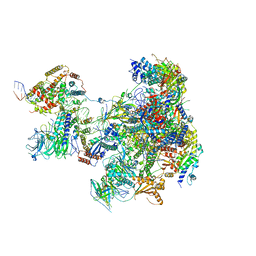 | | RNA Polymerase I Pre-initiation complex DNA opening intermediate 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Mueller, C.W, Sadian, Y, Tafur, L. | | Deposit date: | 2019-05-28 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular insight into RNA polymerase I promoter recognition and promoter melting.
Nat Commun, 10, 2019
|
|
4O1M
 
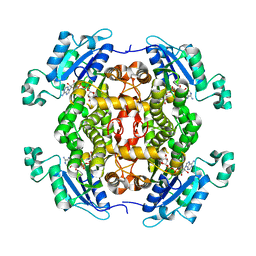 | | Toxoplasma gondii Enoyl acyl carrier protein reductase | | Descriptor: | Enoyl-acyl carrier reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Muench, S.P, Wilkinson, C, Prigge, S.T, Rice, D.W. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The benzimidazole based drugs show good activity against T. gondii but poor activity against its proposed enoyl reductase enzyme target
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1Y1N
 
 | | Identification of SH3 motif in M. Tuberculosis methionine aminopeptidase suggests a mode of interaction with the ribosome | | Descriptor: | Methionine aminopeptidase 1B, POTASSIUM ION | | Authors: | Addlagatta, A, Quillin, M.L, Omotoso, O, Liu, J.O, Matthews, B.W. | | Deposit date: | 2004-11-18 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Identification of an SH3-Binding Motif in a New Class of Methionine Aminopeptidases from Mycobacterium tuberculosis Suggests a Mode of Interaction with the Ribosome
Biochemistry, 44, 2005
|
|
5ELZ
 
 | | Staphylococcus aureus Type II pantothenate kinase in complex with a pantothenate analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, N-(1,3-benzodioxol-5-ylmethyl)-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Mottaghi, K, Hughes, S.J, Tempel, W, Hong, B, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-05 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent Pantothenamide Inhibitors of Staphylococcus aureus Pantothenate Kinase through a Minimal SAR Study: Inhibition Is Due to Trapping of the Product.
Acs Infect Dis., 2, 2016
|
|
