6TX4
 
 | |
4Q3V
 
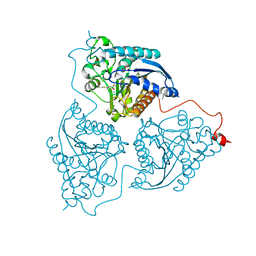 | | Crystal structure of Schistosoma mansoni arginase in complex with inhibitor BEC | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Hai, Y, Edwards, J.E, Van Zandt, M.C, Hoffmann, K.F, Christianson, D.W. | | Deposit date: | 2014-04-12 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Schistosoma mansoni Arginase, a Potential Drug Target for the Treatment of Schistosomiasis.
Biochemistry, 53, 2014
|
|
3IS9
 
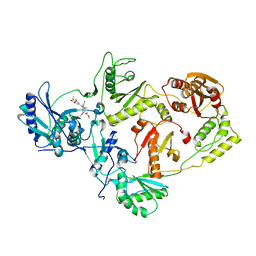 | | Crystal structure of the HIV-1 reverse transcriptase (RT) in complex with the alkenyldiarylmethane (ADAM) Non-nucleoside RT Inhibitor dimethyl 3,3'-(6-methoxy-6-oxohex-1-ene-1,1-diyl)bis(5-cyano-6-methoxybenzoate). | | Descriptor: | Reverse transcriptase, Reverse transcriptase/ribonuclease H, dimethyl 3,3'-(6-methoxy-6-oxohex-1-ene-1,1-diyl)bis(5-cyano-6-methoxybenzoate) | | Authors: | Ho, W.C, Bauman, J.D, Das, K, Arnold, E. | | Deposit date: | 2009-08-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic study of a novel subnanomolar inhibitor provides insight on the binding interactions of alkenyldiarylmethanes with human immunodeficiency virus-1 reverse transcriptase.
J.Med.Chem., 52, 2009
|
|
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
3IJ0
 
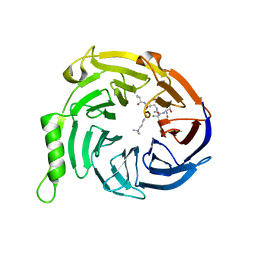 | | Crystal structure of Eed in complex with a trimethylated histone H3K9 peptide | | Descriptor: | Histone H3K9 peptide, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
3IJC
 
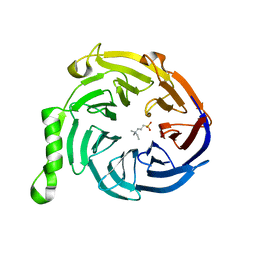 | | Crystal structure of Eed in complex with NDSB-195 | | Descriptor: | ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, Polycomb protein EED | | Authors: | Justin, N, Sharpe, M.L, Martin, S, Taylor, W.R, De Marco, V, Gamblin, S.J. | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of the polycomb protein EED in the propagation of repressive histone marks.
Nature, 461, 2009
|
|
4LKI
 
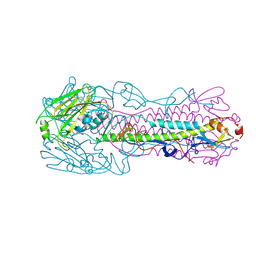 | | The structure of hemagglutinin L226Q mutant from a avian-origin H7N9 influenza virus (A/Anhui/1/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | Authors: | Shi, Y, Zhang, W, Wang, F, Qi, J, Song, H, Wu, Y, Gao, F, Zhang, Y, Fan, Z, Gong, W, Wang, D, Shu, Y, Wang, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-07-07 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses
Science, 342, 2013
|
|
7TXT
 
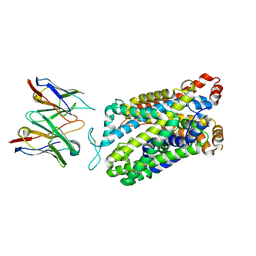 | | Structure of human serotonin transporter bound to small molecule '8090 in lipid nanodisc and NaCl | | Descriptor: | 1-[4-(4-fluorophenyl)-1,3-thiazol-2-yl]piperazine, 15B8 Fab heavy chain, 15B8 Fab light chain, ... | | Authors: | Singh, I, Seth, A, Billesboelle, C.B, Braz, J, Rodriguiz, R.M, Roy, K, Bekele, B, Craik, V, Huang, X.P, Boytsov, D, Lak, P, O'Donnell, H, Sandtner, W, Roth, B.L, Basbaum, A.I, Wetsel, W.C, Manglik, A, Shoichet, B.K, Rudnick, G. | | Deposit date: | 2022-02-09 | | Release date: | 2023-03-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based discovery of conformationally selective inhibitors of the serotonin transporter.
Cell, 186, 2023
|
|
6U4Z
 
 | |
7TTD
 
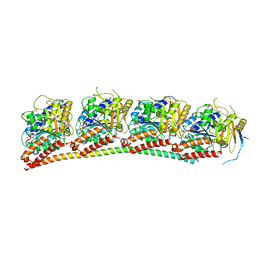 | | Tubulin-RB3_SLD in complex with compound 12e | | Descriptor: | 7-methoxy-4-[2-(morpholin-4-yl)-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl]-3,4-dihydroquinoxalin-2(1H)-one, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2022-02-01 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Pyrimidine Dihydroquinoxalinone Derivatives as Tubulin Colchicine Site-Binding Agents That Displayed Potent Anticancer Activity Both In Vitro and In Vivo.
Acs Pharmacol Transl Sci, 6, 2023
|
|
1L97
 
 | |
7TTE
 
 | | Tubulin-RB3_SLD in complex with compound 12j | | Descriptor: | 4-[2-(cyclopropylamino)-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl]-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2022-02-01 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Pyrimidine Dihydroquinoxalinone Derivatives as Tubulin Colchicine Site-Binding Agents That Displayed Potent Anticancer Activity Both In Vitro and In Vivo.
Acs Pharmacol Transl Sci, 6, 2023
|
|
6U7Z
 
 | | RNA hairpin structure containing one TNA nucleotide as primer | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, RNA (5'-R(*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3'), RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*GP*CP*CP*GP*AP*AP*AP*GP*(TG))-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
3IUF
 
 | | Crystal structure of the C2H2-type zinc finger domain of human ubi-d4 | | Descriptor: | ZINC ION, Zinc finger protein ubi-d4 | | Authors: | Tempel, W, Xu, C, Bian, C, Adams-Cioaba, M, Eryilmaz, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Cys2His2-type zinc finger domain of human DPF2.
Biochem.Biophys.Res.Commun., 413, 2011
|
|
3IUX
 
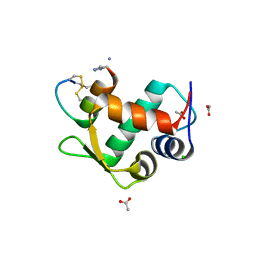 | |
7TXX
 
 | |
2J9D
 
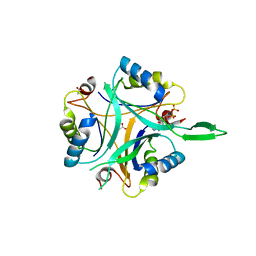 | | Structure of GlnK1 with bound effectors indicates regulatory mechanism for ammonia uptake | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yildiz, O, Kalthoff, C, Raunser, S, Kuehlbrandt, W. | | Deposit date: | 2006-11-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Glnk1 with Bound Effectors Indicates Regulatory Mechanism for Ammonia Uptake.
Embo J., 26, 2007
|
|
5FTY
 
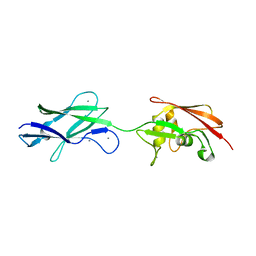 | | Structure of surface layer protein SbsC, domains 6-7 (monoclinic form) | | Descriptor: | CALCIUM ION, SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2016-01-18 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Surface Layer Protein Sbsc, Domains 6-7 (Monoclinic Form)
To be Published
|
|
7KQN
 
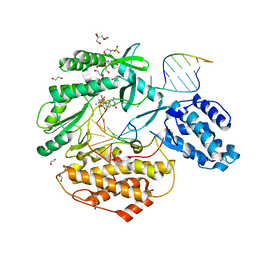 | | Ternary complex of TERT (telomerase reverse transcriptase) with RNA template, DNA primer, an incoming dGTP and a downstream hybrid duplex | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Choi, W.S, Weng, P.J, Yang, W. | | Deposit date: | 2020-11-17 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Flexibility of telomerase in binding the RNA template and DNA telomeric repeat.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U5V
 
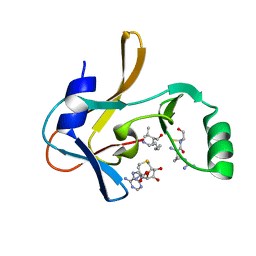 | | Crystal structure of the Mixed Lineage Leukaemia (MLL1) SET Domain with the cofactor product S-Adenosylhomocysteine and Borealin peptide | | Descriptor: | Borealin, Histone-lysine N-methyltransferase 2A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | An, S, Cho, U.S, Oh, H, Sha, L, Xu, J, Kim, S, Yang, W, An, W, Dou, Y. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Non-canonical MLL1 activity regulates centromeric phase separation and genome stability.
Nat.Cell Biol., 25, 2023
|
|
7XK8
 
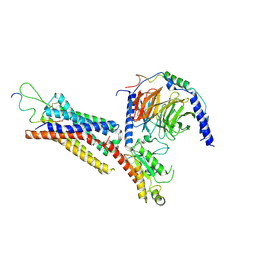 | | Cryo-EM structure of the Neuromedin U receptor 2 (NMUR2) in complex with G Protein and its endogeneous Peptide-Agonist NMU25 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, W, Wenru, Z, Mu, W, Minmin, L, Shutian, C, Tingting, T, Gisela, S, Holger, W, Albert, B, Cuiying, Y, Xiaojing, C, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and activation of neuromedin U receptor 2.
Nat Commun, 13, 2022
|
|
3SZ5
 
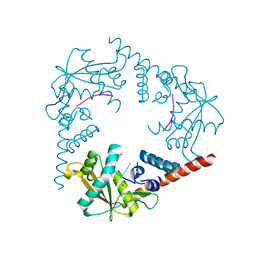 | | Crystal Structure of LHK-Exo in complex with 5-phosphorylated oligothymidine (dT)4 | | Descriptor: | 5'-D(P*TP*TP*TP*T)-3', Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
7UYS
 
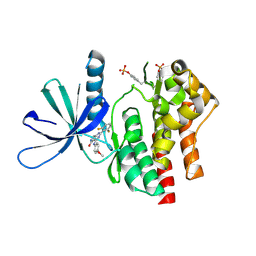 | | Crystal structure of TYK2 kinase domain in complex with compound 16 | | Descriptor: | 2-(2,6-difluorophenyl)-4-(4-methoxyanilino)-5H-pyrrolo[3,4-d]pyrimidin-5-one, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Toms, A.V, Leit, S, Greenwood, J.R, Mondal, S, Carriero, S, Dahlgren, M, Harriman, G.C, Kennedy-Smith, J.J, Kapeller, R, Lawson, J.P, Romero, D.L, Shelley, M, Wester, R.T, Westlin, W, Mc Elwee, J.J, Miao, W, Edmondson, S.D, Massee, C.E. | | Deposit date: | 2022-05-07 | | Release date: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and selective TYK2-JH1 inhibitors highly efficacious in rodent model of psoriasis.
Bioorg.Med.Chem.Lett., 73, 2022
|
|
7CF6
 
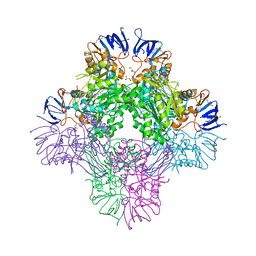 | | Crystal structure of Beta-aspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum AW-1 in complex with beta-Asp-Leu dipeptide | | Descriptor: | (2S)-2-[[(3S)-3-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]amino]-4-methyl-pentanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
1FVG
 
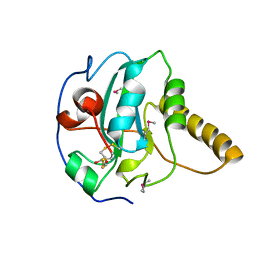 | | CRYSTAL STRUCTURE OF BOVINE PEPTIDE METHIONINE SULFOXIDE REDUCTASE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PEPTIDE METHIONINE SULFOXIDE REDUCTASE | | Authors: | Lowther, W.T, Brot, N, Weissbach, H, Matthews, B.W. | | Deposit date: | 2000-09-19 | | Release date: | 2000-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of peptide methionine sulfoxide reductase, an "anti-oxidation" enzyme.
Biochemistry, 39, 2000
|
|
