2I25
 
 | |
2I27
 
 | |
2I24
 
 | |
2I26
 
 | |
5VGA
 
 | | Alternative model for Fab 36-65 | | Descriptor: | Fab 36-65 heavy chain, Fab 36-65 light chain, TRIETHYLENE GLYCOL | | Authors: | Stanfield, R.L, Rupp, B, Wlodawer, A, Dauter, Z, Porebski, P.J, Minor, W, Jaskolski, M, Pozharski, E, Weichenberger, C.X. | | Deposit date: | 2017-04-10 | | Release date: | 2017-12-06 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5U65
 
 | | Camel Nanobody VHH-5 | | Descriptor: | SULFATE ION, VHH-5 | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers.
Sci Rep, 7, 2017
|
|
5U64
 
 | | Camel nanobody VHH-28 | | Descriptor: | VHH-28 | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (1.153 Å) | | Cite: | Selection of nanobodies with broad neutralizing potential against primary HIV-1 strains using soluble subtype C gp140 envelope trimers.
Sci Rep, 7, 2017
|
|
5T3S
 
 | | HIV gp140 trimer MD39-10MUTA in complex with Fabs PGT124 and 35022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2016-08-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | HIV Vaccine Design to Target Germline Precursors of Glycan-Dependent Broadly Neutralizing Antibodies.
Immunity, 45, 2016
|
|
7LVS
 
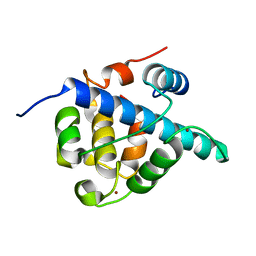 | | The CBP TAZ1 Domain in Complex with a CITED2-HIF-1-Alpha Fusion Peptide | | Descriptor: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone lysine acetyltransferase CREBBP, ZINC ION | | Authors: | Appling, F.D, Berlow, R.B, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The molecular basis of allostery in a facilitated dissociation process.
Structure, 29, 2021
|
|
3K6T
 
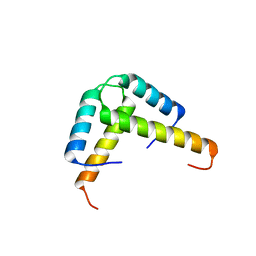 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans at 2.04 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-09 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
3KBL
 
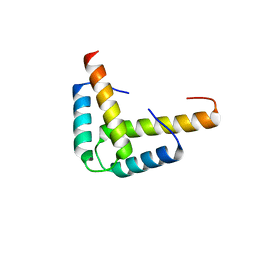 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans N169A mutant at 2.28 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-20 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
6UNA
 
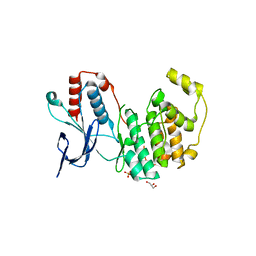 | | Crystal structure of inactive p38gamma | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 12, SULFATE ION | | Authors: | Aoto, P.C, Stanfield, R.L, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2019-10-11 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A Dynamic Switch in Inactive p38 gamma Leads to an Excited State on the Pathway to an Active Kinase.
Biochemistry, 58, 2019
|
|
6V8U
 
 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a modified Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*CP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*GP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | A Conformational Switch in the Zinc Finger Protein Kaiso Mediates Differential Readout of Specific and Methylated DNA Sequences.
Biochemistry, 59, 2020
|
|
3C2A
 
 | | Antibody Fab fragment 447-52D in complex with UG1033 peptide | | Descriptor: | Envelope glycoprotein, Fab 447-52D heavy chain, Fab 447-52D light chain | | Authors: | Dhillon, A.K, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2008-01-24 | | Release date: | 2008-07-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of an anti-HIV-1 Fab 447-52D-peptide complex from an epitaxially twinned data set
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7JPH
 
 | | Crystal structure of EBOV glycoprotein with modified HR1c and HR2 stalk at 3.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, ... | | Authors: | Chaudhary, A, Stanfield, R.L, Wilson, I.A, Zhu, J. | | Deposit date: | 2020-08-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Single-component multilayered self-assembling nanoparticles presenting rationally designed glycoprotein trimers as Ebola virus vaccines.
Nat Commun, 12, 2021
|
|
7JPI
 
 | | Crystal structure of EBOV glycoprotein with modified HR2 stalk at 2.3A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, ... | | Authors: | Chaudhary, A, Stanfield, R.L, Wilson, I.A, Zhu, J. | | Deposit date: | 2020-08-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Single-component multilayered self-assembling nanoparticles presenting rationally designed glycoprotein trimers as Ebola virus vaccines.
Nat Commun, 12, 2021
|
|
6BL6
 
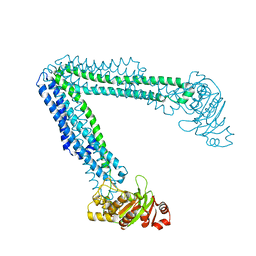 | | Crystallization of lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Stanfield, R.L, Zhang, Q, Wilson, I.A, Lee, S.C. | | Deposit date: | 2017-11-09 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
8T5X
 
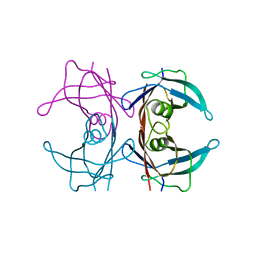 | | Probing the dissociation pathway of a kinetically labile transthyretin mutant (A25T) | | Descriptor: | Transthyretin | | Authors: | Ferguson, J.A, Sun, X, Leach, B.I, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Probing the Dissociation Pathway of a Kinetically Labile Transthyretin Mutant.
J.Am.Chem.Soc., 146, 2024
|
|
7U5G
 
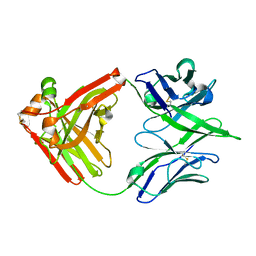 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
7U0K
 
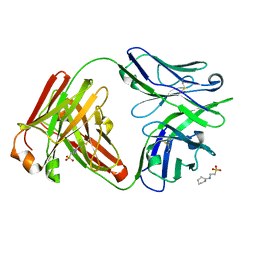 | | IOMA class antibody Fab ACS124 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, IOMA Class antibody ACS124 Heavy chain, IOMA Class antibody ACS124 Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U04
 
 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
2B4C
 
 | | Crystal structure of HIV-1 JR-FL gp120 core protein containing the third variable region (V3) complexed with CD4 and the X5 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, T-cell surface glycoprotein CD4, ... | | Authors: | Huang, C, Tang, M, Zhang, M.Y, Majeed, S, Montabana, E, Stanfield, R.L, Dimitrov, D.S, Korber, B, Sodroski, J, Wilson, I.A, Wyatt, R, Kwong, P.D. | | Deposit date: | 2005-09-23 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a V3-containing HIV-1 gp120 core.
Science, 310, 2005
|
|
1TZG
 
 | | Crystal structure of HIV-1 neutralizing human Fab 4E10 in complex with a 13-residue peptide containing the 4E10 epitope on gp41 | | Descriptor: | Envelope polyprotein GP160, Fab 4E10, GLYCEROL | | Authors: | Cardoso, R.M.F, Zwick, M.B, Stanfield, R.L, Kunert, R, Binley, J.M, Katinger, H, Burton, D.R, Wilson, I.A. | | Deposit date: | 2004-07-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Broadly Neutralizing Anti-HIV Antibody 4E10 Recognizes a Helical Conformation of a Highly Conserved Fusion-Associated Motif in gp41
Immunity, 22, 2005
|
|
1TCR
 
 | | MURINE T-CELL ANTIGEN RECEPTOR 2C CLONE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garcia, K.C, Degano, M, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 1996-09-12 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex.
Science, 274, 1996
|
|
2OQJ
 
 | | Crystal structure analysis of Fab 2G12 in complex with peptide 2G12.1 | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, peptide 2G12.1 (ACPPSHVLDMRSGTCLAAEGK) | | Authors: | Calarese, D.A, Stanfield, R.L, Menendez, A, Scott, J.K, Wilson, I.A. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120.
Faseb J., 22, 2008
|
|
