1V54
 
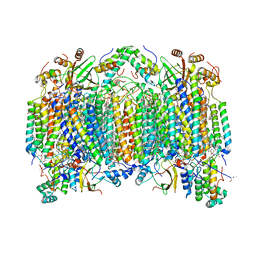 | | Bovine heart cytochrome c oxidase at the fully oxidized state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1V55
 
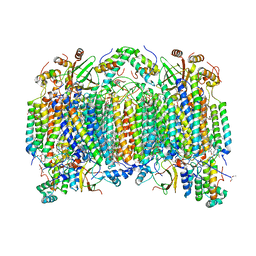 | | Bovine heart cytochrome c oxidase at the fully reduced state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
3A0T
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) in complex with ADP and Mg ion (trigonal) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A0Z
 
 | | Catalytic domain of histidine kinase ThkA (TM1359) (nucleotide free form 4: isopropanol, orthorombic) | | Descriptor: | Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-25 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A0U
 
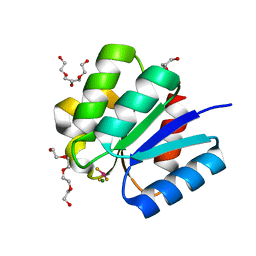 | | Crystal structure of response regulator protein TrrA (TM1360) from Thermotoga maritima in complex with Mg(2+)-BeF (wild type) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator, ... | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A10
 
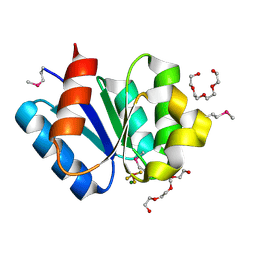 | | Crystal structure of response regulator protein TrrA (TM1360) from Thermotoga maritima in complex with Mg(2+)-BeF (SeMet, L89M) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator, ... | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-25 | | Release date: | 2009-10-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
2E5E
 
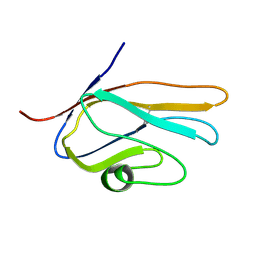 | | Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts | | Descriptor: | Advanced glycosylation end product-specific receptor | | Authors: | Matsumoto, S, Yoshida, T, Yasumatsu, I, Yamamoto, H, Kobayashi, Y, Ohkubo, T. | | Deposit date: | 2006-12-20 | | Release date: | 2007-12-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Variable-type Domain of Human Receptor for Advanced Glycation Endproducts
to be published
|
|
2CWN
 
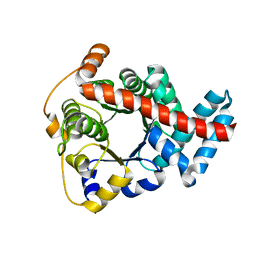 | | Crystal structure of mouse transaldolase | | Descriptor: | Transaldolase | | Authors: | Handa, N, Arai, R, Nishino, A, Uchikubo, T, Takemoto, C, Morita, S, Kinoshita, Y, Nagano, Y, Uda, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of mouse transaldolase
To be Published
|
|
2ZOX
 
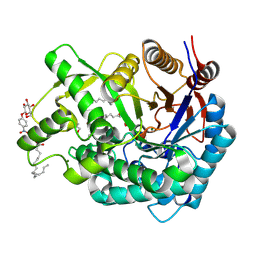 | | Crystal Structure of the Covalent Intermediate of Human Cytosolic beta-Glucosidase | | Descriptor: | 4-nitrophenyl alpha-D-glucopyranoside, Cytosolic beta-glucosidase, GLYCEROL, ... | | Authors: | Noguchi, J, Hayashi, Y, Baba, Y, Okino, N, Kimura, M, Ito, M, Kakuta, Y. | | Deposit date: | 2008-06-17 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the covalent intermediate of human cytosolic beta-glucosidase
Biochem.Biophys.Res.Commun., 374, 2008
|
|
1X3U
 
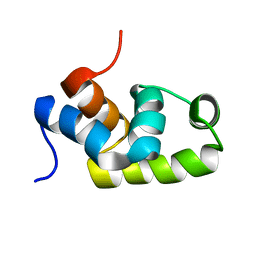 | | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium melilot | | Descriptor: | Transcriptional regulatory protein fixJ | | Authors: | Kurashima-Ito, K, Kasai, Y, Hosono, K, Tamura, K, Oue, S, Isogai, M, Ito, Y, Nakamura, H, Shiro, Y. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium meliloti and its recognition of the fixK promoter
Biochemistry, 44, 2005
|
|
2E33
 
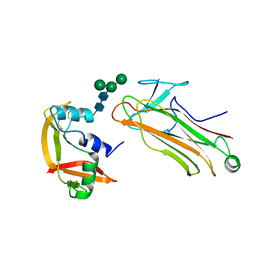 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, Ribonuclease pancreatic, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E31
 
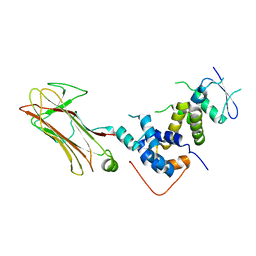 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, S-phase kinase-associated protein 1A | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2E2R
 
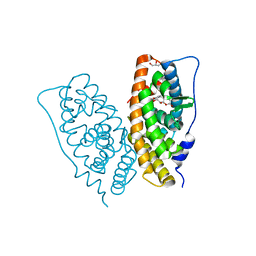 | | Crystal structure of human estrogen-related receptor gamma ligand binding domain complex with bisphenol A | | Descriptor: | 4,4'-PROPANE-2,2-DIYLDIPHENOL, Estrogen-related receptor gamma, GLYCEROL | | Authors: | Matsushima, A, Kakuta, Y, Teramoto, T, Koshiba, T, Kimura, M, Shimohigashi, Y. | | Deposit date: | 2006-11-16 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Evidence for Endocrine Disruptor Bisphenol A Binding to Human Nuclear Receptor ERR{gamma}
J.Biochem.(Tokyo), 142, 2007
|
|
2E32
 
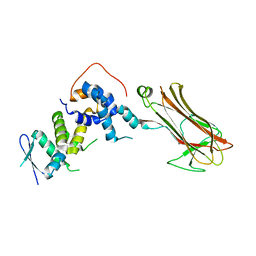 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, S-phase kinase-associated protein 1A | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2CW9
 
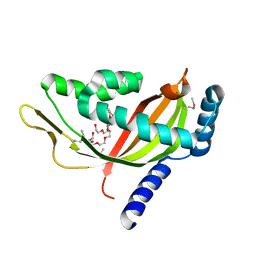 | | Crystal structure of human Tim44 C-terminal domain | | Descriptor: | PENTAETHYLENE GLYCOL, translocase of inner mitochondrial membrane | | Authors: | Handa, N, Kishishita, S, Morita, S, Kinoshita, Y, Nagano, Y, Uda, H, Terada, T, Uchikubo, T, Takemoto, C, Jin, Z, Chrzas, J, Chen, L, Liu, Z.-J, Wang, B.-C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-17 | | Release date: | 2005-12-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human Tim44 C-terminal domain in complex with pentaethylene glycol: ligand-bound form.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2D6C
 
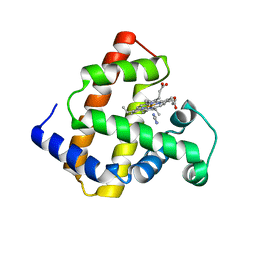 | | Crystal structure of myoglobin reconstituted with iron porphycene | | Descriptor: | IMIDAZOLE, Myoglobin, PORPHYCENE CONTAINING FE | | Authors: | Hayashi, T, Murata, D, Makino, M, Sugimoto, H, Matsuo, T, Sato, H, Shiro, Y, Hisaeda, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-11 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure and peroxidase activity of myoglobin reconstituted with iron porphycene
Inorg.Chem., 45, 2006
|
|
2DBQ
 
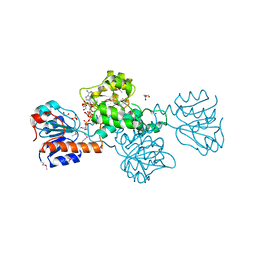 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (I41) | | Descriptor: | GLYCEROL, Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2DBZ
 
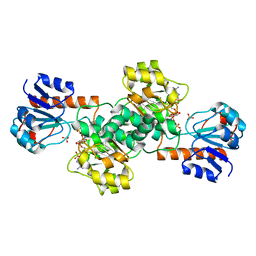 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (P61) | | Descriptor: | Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2DBR
 
 | | Crystal Structure of Glyoxylate Reductase (PH0597) from Pyrococcus horikoshii OT3, Complexed with NADP (P1) | | Descriptor: | Glyoxylate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kinoshita, Y, Uchikubo-Kamo, T, Akasaka, R, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-16 | | Release date: | 2006-06-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of archaeal glyoxylate reductase from Pyrococcus horikoshii OT3 complexed with nicotinamide adenine dinucleotide phosphate.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2EKT
 
 | | Crystal structure of myoglobin reconstituted with 6-methyl-6-depropionatehemin | | Descriptor: | 6-METHY-6-DEPROPIONATEHEMIN, Myoglobin, SULFATE ION | | Authors: | Harada, K, Makino, M, Sugimoto, H, Hirota, S, Matsuo, T, Shiro, Y, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2007-03-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and ligand binding properties of myoglobins reconstituted with monodepropionated heme: functional role of each heme propionate side chain
Biochemistry, 46, 2007
|
|
2EKU
 
 | | Crystal structure of myoglobin reconstituted with 7-methyl-7-depropionatehemin | | Descriptor: | 7-METHYL-7-DEPROPIONATEHEMIN, Myoglobin, SULFATE ION | | Authors: | Harada, K, Makino, M, Sugimoto, H, Hirota, S, Matsuo, T, Shiro, Y, Hisaeda, Y, Hayashi, T. | | Deposit date: | 2007-03-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and ligand binding properties of myoglobins reconstituted with monodepropionated heme: functional role of each heme propionate side chain
Biochemistry, 46, 2007
|
|
3CVF
 
 | | Crystal Structure of the carboxy terminus of Homer3 | | Descriptor: | Homer protein homolog 3 | | Authors: | Hayashi, M.K, Stearns, M.H, Giannini, V, Xu, R.-M, Sala, C, Hayashi, Y. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The postsynaptic density proteins Homer and Shank form a polymeric network structure.
Cell(Cambridge,Mass.), 137, 2009
|
|
3CVE
 
 | | Crystal Structure of the carboxy terminus of Homer1 | | Descriptor: | Homer protein homolog 1 | | Authors: | Hayashi, M.K, Stearns, M.H, Giannini, V, Xu, R.-M, Sala, C, Hayashi, Y. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-31 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The postsynaptic density proteins Homer and Shank form a polymeric network structure.
Cell(Cambridge,Mass.), 137, 2009
|
|
