1J3Y
 
 | | Direct observation of photolysis-induced tertiary structural changes in human hemoglobin; Crystal structure of alpha(Fe)-beta(Ni) hemoglobin (laser photolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1IZ5
 
 | | Pyrococcus furiosus PCNA mutant (Met73Leu, Asp143Ala, Asp147Ala): orthorhombic form | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Matsumiya, S, Ishino, S, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-09-23 | | Release date: | 2003-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intermolecular ion pairs maintain the toroidal structure of Pyrococcus furiosus PCNA
PROTEIN SCI., 12, 2003
|
|
1WR6
 
 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
5YUA
 
 | | Crystal structure of voltage-gated sodium channel NavAb in high-pH condition | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHAPSO, ... | | Authors: | Irie, K, Shimomura, T, Fujiyoshi, Y. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.80085564 Å) | | Cite: | Structural insight on the voltage dependence of prokaryotic voltage gated sodium channel NavAb.
FEBS Lett., 2017
|
|
5YUC
 
 | |
5ZLG
 
 | | Human duodenal cytochrome b (Dcytb) in zinc ion and ascorbate bound form | | Descriptor: | ASCORBIC ACID, Cytochrome b reductase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ganasen, M, Togashi, H, Mauk, G.A, Shiro, Y, Sawai, H, Sugimoto, H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate.
Commun Biol, 1, 2018
|
|
3VM4
 
 | | Cytochrome P450SP alpha (CYP152B1) in complex with (R)-ibuprophen | | Descriptor: | (2R)-2-[4-(2-methylpropyl)phenyl]propanoic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Chiral-substrate-assisted stereoselective epoxidation catalyzed by H2O2-dependent cytochrome P450SP alpha
Chem Asian J, 7, 2012
|
|
3VQI
 
 | | Crystal structure of Kluyveromyces marxianus Atg5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Atg5, SULFATE ION | | Authors: | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-03-24 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
7X8V
 
 | | Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity | | Descriptor: | Os01g0156300 protein | | Authors: | Ichimaru, K, Harada, K, Yamaguchi, K, Shigeta, S, Shimada, K, Ishikawa, K, Inoue, K, Nishio, Y, Yoshimura, S, Inoue, H, Yamashita, E, Fujiwara, T, Nakagawa, A, Kojima, C, Kawasaki, T. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Cooperative regulation of PBI1 and MAPKs controls WRKY45 transcription factor in rice immunity.
Nat Commun, 13, 2022
|
|
3W8M
 
 | | Crystal Structure of HasAp with Iron Salophen | | Descriptor: | 2,2'-[1,2-PHENYLENEBIS(NITRILOMETHYLIDYNE)]BIS[PHENOLATO]](2-)-N,N',O,O']-IRON, Heme acquisition protein HasAp | | Authors: | Shirataki, C, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2013-03-19 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Inhibition of Heme Uptake in Pseudomonas aeruginosa by its Hemophore (HasAp ) Bound to Synthetic Metal Complexes
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3W8O
 
 | | Crystal Structure of HasAp with Iron Phthalocyanine | | Descriptor: | Heme acquisition protein HasAp, [29H,31H-phthalocyaninato(2-)-kappa~4~N~29~,N~30~,N~31~,N~32~]iron | | Authors: | Shirataki, C, Shoji, O, Terada, M, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2013-03-20 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inhibition of Heme Uptake in Pseudomonas aeruginosa by its Hemophore (HasAp ) Bound to Synthetic Metal Complexes
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8JS7
 
 | | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme | | Descriptor: | GLYCEROL, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kamaya, M, Koteishi, H, Sawai, H, Sugimoto, H, Shiro, Y. | | Deposit date: | 2023-06-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme.
To be published
|
|
8JS5
 
 | | Dimeric PAS domains of oxygen sensor FixL with ferric unliganded heme | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Sensor protein FixL | | Authors: | Kamaya, M, Koteishi, H, Sawai, H, Sugimoto, H, Shiro, Y. | | Deposit date: | 2023-06-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme.
To be published
|
|
8JS6
 
 | | Dimeric PAS domains of oxygen sensor FixL in complex with cyanide-bound ferric heme | | Descriptor: | CYANIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kamaya, M, Koteishi, H, Sawai, H, Sugimoto, H, Shiro, Y. | | Deposit date: | 2023-06-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme.
To be published
|
|
5H1R
 
 | |
5GJ3
 
 | | Periplasmic heme-binding protein RhuT from Roseiflexus sp. RS-1 in two-heme bound form (holo-2) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Periplasmic binding protein, ZINC ION | | Authors: | Rahman, M.M, Naoe, Y, Nakamura, N, Shiro, Y, Sugimoto, H. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems.
Proteins, 85, 2017
|
|
5GIZ
 
 | | Periplasmic heme-binding protein BhuT in apo form | | Descriptor: | CHLORIDE ION, Putative hemin transport system, substrate-binding protein, ... | | Authors: | Nakamura, N, Naoe, Y, Rahman, M.M, Shiro, Y, Sugimoto, H. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems.
Proteins, 85, 2017
|
|
7WGO
 
 | | X-ray structure of human PPAR gamma ligand binding domain-bezafibrate co-rystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGP
 
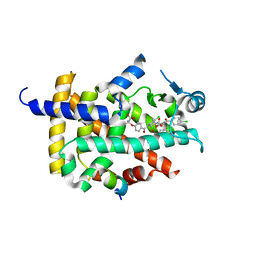 | | X-ray structure of human PPAR gamma ligand binding domain-fenofibric acid co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGQ
 
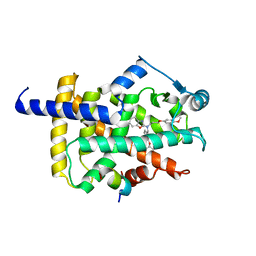 | | X-ray structure of human PPAR gamma ligand binding domain-pemafibrate co-crystals obtained by co-crystallization | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, 15-meric peptide from Nuclear receptor coactivator 1, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGN
 
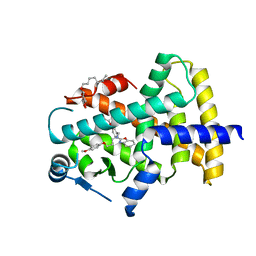 | | X-ray structure of human PPAR delta ligand binding domain-pemafibrate co-crystals obtained by co-crystallization | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7WGL
 
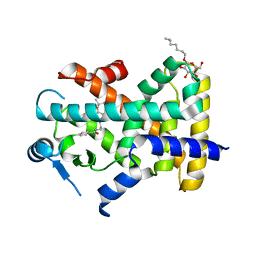 | | X-ray structure of human PPAR delta ligand binding domain-bezafibrate co-crystals obtained by co-crystallization | | Descriptor: | 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
5H1Q
 
 | |
2PCR
 
 | | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5 | | Descriptor: | Inositol-1-monophosphatase | | Authors: | Jeyakanthan, J, Gayathri, D, Velmurugan, D, Agari, Y, Bessho, Y, Ellis, M.J, Antonyuk, S.V, Strange, R.W, Hasnain, S.S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5
To be Published
|
|
5ZLE
 
 | | Human duodenal cytochrome b (Dcytb) in substrate free form | | Descriptor: | Cytochrome b reductase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ganasen, M, Togashi, H, Mauk, G.A, Shiro, Y, Sawai, H, Sugimoto, H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate.
Commun Biol, 1, 2018
|
|
