6OZ1
 
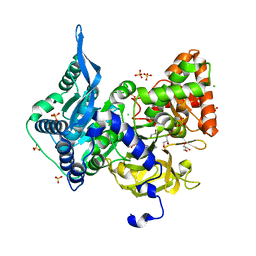 | | Crystal structure of the adenylation (A) domain of the carboxylate reductase (CAR) GR01_22995 from Mycobacterium chelonae | | 分子名称: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Evdokimova, E, Di Leo, R, Fedorchuk, T, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | 登録日 | 2019-05-15 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | One-Pot Biocatalytic Transformation of Adipic Acid to 6-Aminocaproic Acid and 1,6-Hexamethylenediamine Using Carboxylic Acid Reductases and Transaminases.
J.Am.Chem.Soc., 142, 2020
|
|
6P2L
 
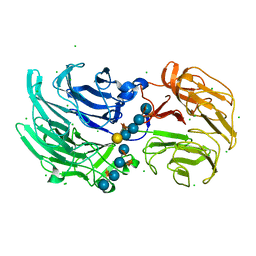 | | Crystal structure of Niastella koreensis GH74 (NkGH74) enzyme | | 分子名称: | CHLORIDE ION, Glycosyl hydrolase BNR repeat-containing protein, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | 著者 | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | 登録日 | 2019-05-21 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
1NI9
 
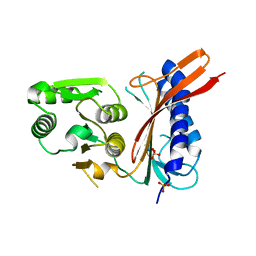 | | 2.0 A structure of glycerol metabolism protein from E. coli | | 分子名称: | Protein glpX, SULFATE ION | | 著者 | Sanishvili, R, Brunzelle, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-12-23 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
1O8B
 
 | | Structure of Escherichia coli ribose-5-phosphate isomerase, RpiA, complexed with arabinose-5-phosphate. | | 分子名称: | 5-O-phosphono-beta-D-arabinofuranose, RIBOSE 5-PHOSPHATE ISOMERASE | | 著者 | Zhang, R.-g, Andersson, C.E, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Mowbray, S.L, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-11-26 | | 公開日 | 2003-01-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structure of Escherichia Coli Ribose-5-Phosphate Isomerase: A Ubiquitous Enzyme of the Pentose Phosphate Pathway and the Calvin Cycle
Structure, 11, 2003
|
|
1NN4
 
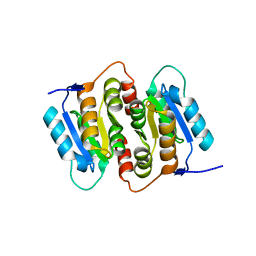 | | Structural Genomics, RpiB/AlsB | | 分子名称: | Ribose 5-phosphate isomerase B | | 著者 | Zhang, R.G, Andersson, C.E, Mowbray, S.L, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S.L, Arrowsmith, C, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-01-12 | | 公開日 | 2003-07-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction.
J.Mol.Biol., 332, 2003
|
|
3VCX
 
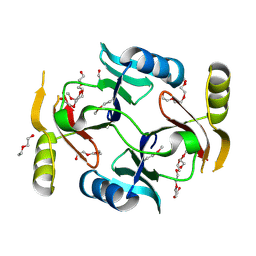 | | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009 | | 分子名称: | Glyoxalase/Bleomycin resistance protein/dioxygenase domain, TETRAETHYLENE GLYCOL | | 著者 | Stogios, P.J, Chang, C, Evdokimova, E, Egorova, O, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-01-04 | | 公開日 | 2012-01-18 | | 最終更新日 | 2012-01-25 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Crystal structure of a putative glyoxalase/bleomycin resistance protein from Rhodopseudomonas palustris CGA009
To be Published
|
|
7TBU
 
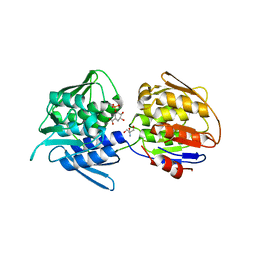 | | Crystal structure of the 5-enolpyruvate-shikimate-3-phosphate synthase (EPSPS) domain of Aro1 from Candida albicans in complex with shikimate-3-phosphate | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-enolpyruvylshikimate-3-phosphate synthase, SHIKIMATE-3-PHOSPHATE | | 著者 | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-12-22 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
7TBV
 
 | | Crystal structure of the shikimate kinase + 3-dehydroquinate dehydratase + 3-dehydroshikimate dehydrogenase domains of Aro1 from Candida albicans | | 分子名称: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-12-22 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
7SNU
 
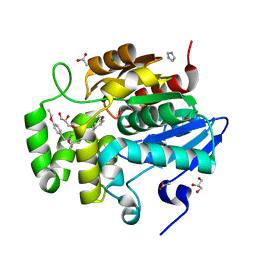 | | Crystal structure of ShHTL7 from Striga hermonthica in complex with strigolactone antagonist RG6 | | 分子名称: | 2-{(2S)-1-[(4-ethoxyphenyl)methyl]-4-[(2E)-3-(4-methoxyphenyl)prop-2-en-1-yl]piperazin-2-yl}ethan-1-ol, ACETATE ION, GLYCEROL, ... | | 著者 | Arellano-Saab, A, Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, McCourt, P. | | 登録日 | 2021-10-28 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | A novel strigolactone receptor antagonist provides insights into the structural inhibition, conditioning, and germination of the crop parasite Striga.
J.Biol.Chem., 298, 2022
|
|
5HMN
 
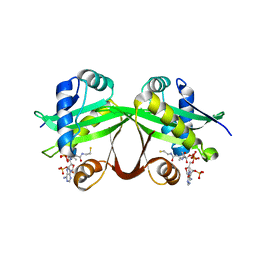 | | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol | | 分子名称: | AAC3-I, COENZYME A, TETRAETHYLENE GLYCOL | | 著者 | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-01-16 | | 公開日 | 2016-02-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.018 Å) | | 主引用文献 | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol
To Be Published
|
|
1NR9
 
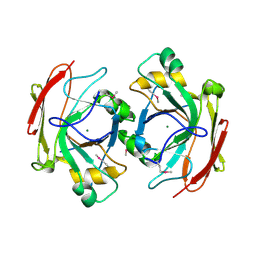 | | Crystal Structure of Escherichia coli 1262 (APC5008), Putative Isomerase | | 分子名称: | MAGNESIUM ION, Protein YCGM | | 著者 | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-01-24 | | 公開日 | 2003-07-29 | | 最終更新日 | 2023-02-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of Escherichia coli Putative Isomerase EC1262 (APC5008)
To be Published
|
|
5SUJ
 
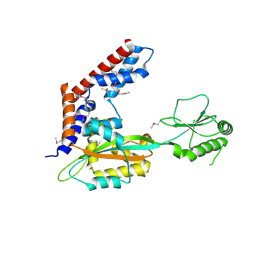 | | Crystal structure of uncharacterized protein LPG2148 from Legionella pneumophila | | 分子名称: | Uncharacterized protein | | 著者 | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2016-08-03 | | 公開日 | 2016-08-17 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.356 Å) | | 主引用文献 | Discovery of Ubiquitin Deamidases in the Pathogenic Arsenal of Legionella pneumophila.
Cell Rep, 23, 2018
|
|
6OZ7
 
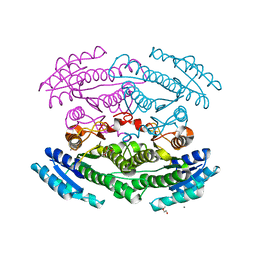 | | Putative oxidoreductase from Escherichia coli str. K-12 | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, TRIETHYLENE GLYCOL, ... | | 著者 | Osipiuk, J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-05-15 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Putative oxidoreductase from Escherichia coli str. K-12
to be published
|
|
1NQK
 
 | | Structural Genomics, Crystal structure of Alkanesulfonate monooxygenase | | 分子名称: | Alkanesulfonate monooxygenase | | 著者 | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-01-21 | | 公開日 | 2003-09-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure of the protein
Alkanesulfonate monooxygenase from E. Coli
To be Published
|
|
8SDC
 
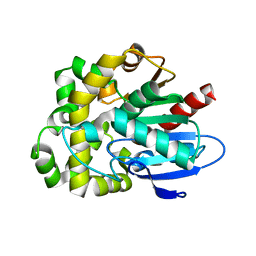 | | Crystal structure of fluoroacetate dehalogenase Daro3835 apoenzyme | | 分子名称: | Alpha/beta hydrolase fold protein, CHLORIDE ION | | 著者 | Stogios, P.J, Skarina, T, Khusnutdinova, A, Iakounine, A, Savchenko, A. | | 登録日 | 2023-04-06 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural insights into hydrolytic defluorination of difluoroacetate by microbial fluoroacetate dehalogenases.
Febs J., 290, 2023
|
|
8SDD
 
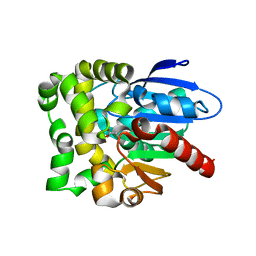 | | Crystal structure of fluoroacetate dehalogenase Daro3835 H274N mutant with D107-glycolyl intermediate | | 分子名称: | Alpha/beta hydrolase fold protein | | 著者 | Stogios, P.J, Skarina, T, Khusnutdinova, A, Iakounine, A, Savchenko, A. | | 登録日 | 2023-04-06 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into hydrolytic defluorination of difluoroacetate by microbial fluoroacetate dehalogenases.
Febs J., 290, 2023
|
|
1NXH
 
 | | X-RAY STRUCTURE: NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT87 | | 分子名称: | MTH396 protein | | 著者 | Khayat, R, Savchenko, A, Edwards, A, Arowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-02-10 | | 公開日 | 2004-02-24 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | X-RAY STRUCTURE OF MTH396
To be Published
|
|
8SCD
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with reaction intermediate | | 分子名称: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, 4-AMINOBENZOIC ACID, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2023-04-05 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
6D2X
 
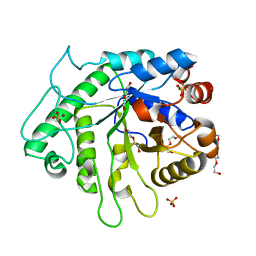 | | Crystal structure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aryl-phospho-beta-D-glucosidase BglC, GH1 family, ... | | 著者 | Stogios, P.J, Skarina, T, McGregor, N, Di Leo, R, Brumer, H, Savchenko, A. | | 登録日 | 2018-04-14 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Crystal structure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii
To Be Published
|
|
3DCI
 
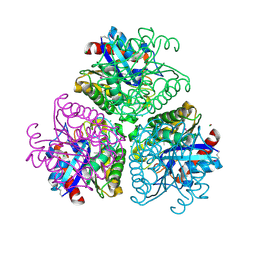 | | The Structure of a putative arylesterase from Agrobacterium tumefaciens str. C58 | | 分子名称: | ACETIC ACID, Arylesterase, CHLORIDE ION, ... | | 著者 | Cuff, M.E, Xu, X, Zheng, H, Binkowski, T.A, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-06-03 | | 公開日 | 2008-09-30 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Structure of a putative arylesterase from Agrobacterium tumefaciens str. C58
TO BE PUBLISHED
|
|
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | 分子名称: | GLYCEROL, putative isochorismatase hydrolase | | 著者 | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-10 | | 公開日 | 2010-03-16 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
3CNG
 
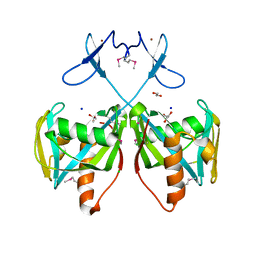 | | Crystal structure of NUDIX hydrolase from Nitrosomonas europaea | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-03-25 | | 公開日 | 2008-04-08 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray crystal structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
3DCL
 
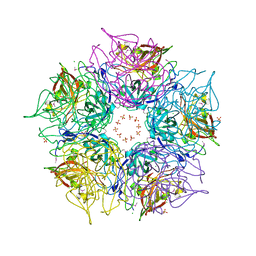 | | Crystal structure of TM1086 | | 分子名称: | CHLORIDE ION, POTASSIUM ION, SULFATE ION, ... | | 著者 | Chruszcz, M, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-06-03 | | 公開日 | 2008-08-05 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal structure of TM1086
To be Published
|
|
3DCA
 
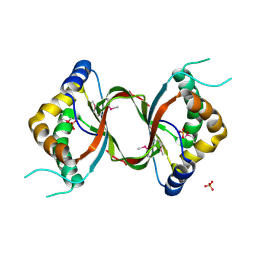 | | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target | | 分子名称: | RPA0582, SULFATE ION | | 著者 | Sledz, P, Wang, S, Chruszcz, M, Yim, V, Kudritska, M, Evdokimova, E, Turk, D, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-06-03 | | 公開日 | 2008-08-05 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Crystal structure of the RPA0582- protein of unknown function from Rhodopseudomonas palustris- a structural genomics target
To be Published
|
|
3D1P
 
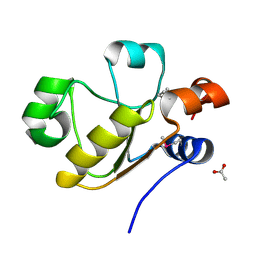 | | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae | | 分子名称: | ACETATE ION, CHLORIDE ION, Putative thiosulfate sulfurtransferase YOR285W | | 著者 | Nocek, B, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-05-06 | | 公開日 | 2008-07-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Atomic resolution structure of uncharacterized protein from Saccharomyces cerevisiae.
To be Published
|
|
