3T3K
 
 | |
5TMN
 
 | | Slow-and fast-binding inhibitors of thermolysin display different modes of binding. crystallographic analysis of extended phosphonamidate transition-state analogues | | Descriptor: | CALCIUM ION, N-[(S)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-L-leucyl-L-leucine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
3T3L
 
 | |
1ZXC
 
 | | Crystal structure of catalytic domain of TNF-alpha converting enzyme (TACE) with inhibitor | | Descriptor: | (3S)-4-{[4-(BUT-2-YNYLOXY)PHENYL]SULFONYL}-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | Authors: | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Zhang, Y, Jin, G, Cowling, R, Barone, D, Skotnicki, J.S. | | Deposit date: | 2005-06-07 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylenic TACE inhibitors. Part 2: SAR of six-membered cyclic sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8AXP
 
 | |
4BQG
 
 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-(3,4-dichloro-phenoxy)-benzene-1,3-diol, HSP90AA1 PROTEIN | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
3F7L
 
 | | X-ray Crystal Structure of Alvinella pompejana Cu,Zn Superoxide Dismutase | | Descriptor: | ACETIC ACID, COPPER (I) ION, COPPER (II) ION, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
4AJD
 
 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | 2-ETHYL-4-METHYL-PHTHALAZIN-1-ONE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
4AJF
 
 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, MAGNESIUM ION, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
4AJM
 
 | | Development of a plate-based optical biosensor methodology to identify PDE10 fragment inhibitors | | Descriptor: | 3-AMINO-6-FLUORO-2-[4-(2-METHYLPYRIDIN-4-YL)PHENYL]-N-(METHYLSULFONYL)QUINOLINE-4-CARBOXAMIDE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Geschwindner, S, Johansson, P, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Albert, J.S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of a Plate-Based Optical Biosensor Methodology to Identify Pde10 Fragment Inhibitors
To be Published
|
|
3F7K
 
 | | X-ray Crystal Structure of an Alvinella pompejana Cu,Zn Superoxide Dismutase- Hydrogen Peroxide Complex | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Copper,Zinc Superoxide Dismutase, ... | | Authors: | Shin, D.S, DiDonato, M, Barondeau, D.P, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Superoxide Dismutase from the Eukaryotic Thermophile Alvinella pompejana: Structures, Stability, Mechanism, and Insights into Amyotrophic Lateral Sclerosis.
J.Mol.Biol., 385, 2009
|
|
5USR
 
 | |
4BQJ
 
 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-[2,4-dihydroxy-6-(4-nitrophenoxy)phenyl]-N-ethyl-1,2-oxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
4AJG
 
 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, MAGNESIUM ION, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
3ITC
 
 | | Crystal structure of Sco3058 with bound citrate and glycerol | | Descriptor: | CITRIC ACID, GLYCEROL, ZINC ION, ... | | Authors: | Nguyen, T.T, Cummings, J.A, Tsai, C.-L, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure, mechanism, and substrate profile for Sco3058: the closest bacterial homologue to human renal dipeptidase
Biochemistry, 49, 2010
|
|
8UM3
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 6-chlorotetrazolo[1,5-b]pyridazine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992
To Be Published
|
|
8V7R
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132 | | Descriptor: | (5R)-5-[2-(4-methoxyphenyl)ethyl]-5-methylimidazolidine-2,4-dione, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132
To Be Published
|
|
8V7U
 
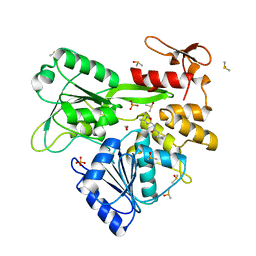 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784
To Be Published
|
|
5S72
 
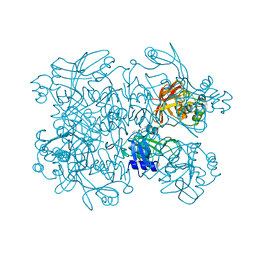 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with BBL029427 | | Descriptor: | CITRIC ACID, N-(2-aminoethyl)-N'-phenylurea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S6Y
 
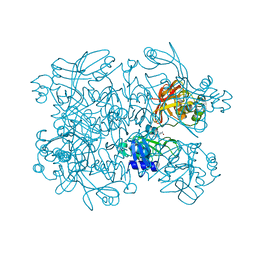 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z56900771 | | Descriptor: | CITRIC ACID, N-[(furan-2-yl)methyl]urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S6Z
 
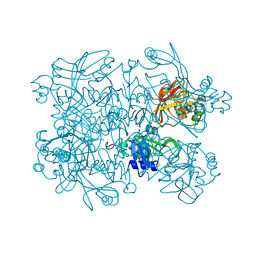 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with PB2255187532 | | Descriptor: | 4-[(dimethylamino)methyl]-1,3-thiazol-2-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S71
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with FUZS-5 | | Descriptor: | 5'-thiothymidine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Bege, M, Bajusz, D, BorbAs, A, Keseru, G.M, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SBF
 
 | | PanDDA analysis group deposition of ground-state model of SARS-CoV-2 NendoU | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-07-23 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
5S6X
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2889976755 | | Descriptor: | 1-(2,4-dimethyl-1H-imidazol-5-yl)methanamine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S70
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-181428 | | Descriptor: | (5R)-2-methyl-4,5,6,7-tetrahydro-1H-benzimidazol-5-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
