3MTV
 
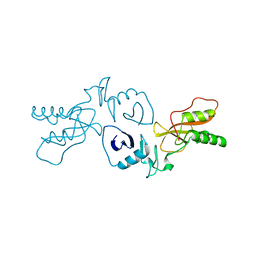 | | The Crystal Structure of the PRRSV Nonstructural Protein Nsp1 | | Descriptor: | Papain-like cysteine protease | | Authors: | Xue, F, Sun, Y.N, Yan, L.M, Zhao, C, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of porcine reproductive and respiratory syndrome virus nonstructural protein Nsp1beta reveals a novel metal-dependent nuclease
J.Virol., 84, 2010
|
|
7ZL9
 
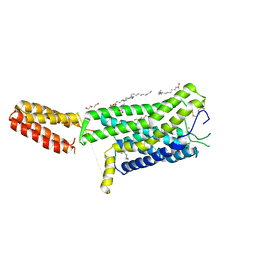 | | Crystal structure of human GPCR Niacin receptor (HCA2) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, FiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7ZLY
 
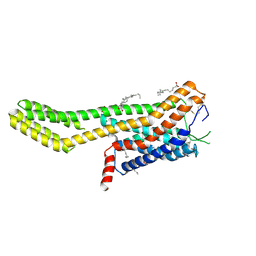 | | Crystal structure of human GPCR Niacin receptor (HCA2) expressed from Spodoptera frugiperda | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, OLEIC ACID | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, FiBerto, J.F, Wu, L.J, Tong, J.H, Han, G.W, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
6J17
 
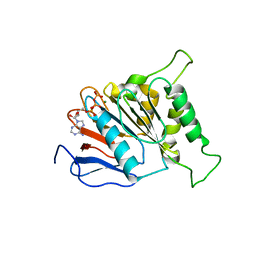 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ESX-3 secretion system protein EccC3, MAGNESIUM ION | | Authors: | Wang, S.H, Li, J, Rao, Z.H. | | Deposit date: | 2018-12-28 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Structural insights into substrate recognition by the type VII secretion system.
Protein Cell, 11, 2020
|
|
6J72
 
 | | Crystal structure of IniA from Mycobacterium smegmatis with GTP bound | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Isoniazid inducible gene protein IniA, L(+)-TARTARIC ACID, ... | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
6JD4
 
 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ESX-1 secretion system protein EccCb1, MAGNESIUM ION | | Authors: | Wang, S.H, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-31 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into substrate recognition by the type VII secretion system.
Protein Cell, 11, 2020
|
|
6J73
 
 | | Crystal structure of IniA from Mycobacterium smegmatis | | Descriptor: | Isoniazid inducible gene protein IniA | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
6JHR
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F6 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHQ
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F4 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHT
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F9 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
6JHS
 
 | | The cryo-EM structure of HAV bound to a neutralizing antibody-F7 | | Descriptor: | FAB Heavy Chain, FAB Light Chain, VP1, ... | | Authors: | Cao, L, Liu, P, Yang, P, Gao, Q, Li, H, Sun, Y, Zhu, L, Lin, J, Su, D, Rao, Z, Wang, X. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors.
Plos Biol., 17, 2019
|
|
5ABJ
 
 | | Structure of Coxsackievirus A16 in complex with GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Tijsma, A, Neyts, J, Spyrou, J.A.B, Ren, J, Grimes, J.M, Puerstinger, G, Leyssen, P, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure Elucidation of Coxsackievirus A16 in Complex with Gpp3 Informs a Systematic Review of Highly Potent Capsid Binders to Enteroviruses.
Plos Pathog., 11, 2015
|
|
4Z9P
 
 | | Crystal structure of Ebola virus nucleoprotein core domain at 1.8A resolution | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Dong, S.S, Yang, P, Li, G.B, Liu, B.C, Yang, C, Rao, Z.H. | | Deposit date: | 2015-04-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution.
Protein Cell, 6, 2015
|
|
8GX3
 
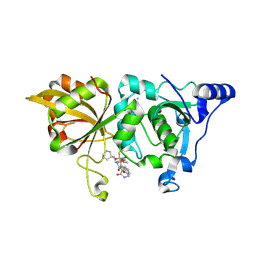 | | The crystal structure of human Calpain-1 protease core in complex with 14c | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14c
To Be Published
|
|
8GX2
 
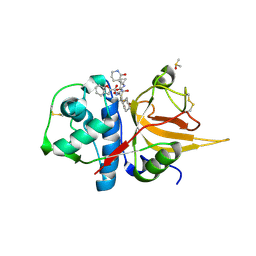 | | The crystal structure of human CtsL in complex with 14c | | Descriptor: | DIMETHYL SULFOXIDE, N-[(2S)-3-cyclohexyl-1-[[(2S,3S)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2022-09-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human CtsL in complex with 14a
To Be Published
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8HCZ
 
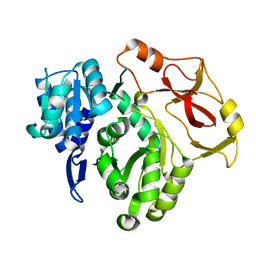 | |
8HDF
 
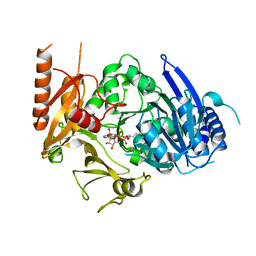 | | Full length crystal structure of mycobacterium tuberculosis FadD23 in complex with ANP and PLM | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, PALMITIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
8HD4
 
 | | Full-length crystal structure of mycobacterium tuberculosis FadD23 in complex with AMPC16 | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, palmitoyl adenylate | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
8GXI
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with 14c | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-3-cyclohexyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-1-benzofuran-2-carboxamide | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of SARS-CoV-2 main protease in complex with 14c
To Be Published
|
|
8WDB
 
 | | Cryo-EM structure of the ATP-bound DppABCD complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable dipeptide-transport ATP-binding protein ABC transporter DppD, ... | | Authors: | Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-09-14 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Cryo-EM structure of the ATP-bound DppABCD complex
To Be Published
|
|
8WD9
 
 | | Cryo-EM structure of Mycobacterium tuberculosis DppABCD in apo form | | Descriptor: | (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, PALMITIC ACID, Probable dipeptide-transport ATP-binding protein ABC transporter DppD, ... | | Authors: | Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-09-14 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of Mycobacterium tuberculosis DppABCD in apo form
To Be Published
|
|
8XFC
 
 | |
