2MFN
 
 | | SOLUTION NMR STRUCTURE OF LINKED CELL ATTACHMENT MODULES OF MOUSE FIBRONECTIN CONTAINING THE RGD AND SYNERGY REGIONS, 10 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Copie, V, Tomita, Y, Akiyama, S.K, Aota, S, Yamada, K.M, Venable, R.M, Pastor, R.W, Krueger, S, Torchia, D.A. | | Deposit date: | 1998-02-11 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure.
J.Mol.Biol., 277, 1998
|
|
1JRT
 
 | |
1JRS
 
 | |
2NST
 
 | | Crystal structure of pectin methylesterase D178A mutant in complex with hexasaccharide II | | Descriptor: | Pectinesterase A, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-06 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
2NTP
 
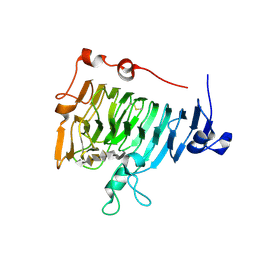 | | Crystal structure of pectin methylesterase in complex with hexasaccharide VI | | Descriptor: | Pectinesterase A, methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, methyl beta-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-08 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
1JMJ
 
 | | Crystal Structure of Native Heparin Cofactor II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEPARIN COFACTOR II, ... | | Authors: | Baglin, T.P, Carrell, R.W, Church, F.C, Huntington, J.A. | | Deposit date: | 2001-07-18 | | Release date: | 2002-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2NTB
 
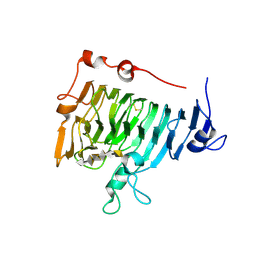 | | Crystal structure of pectin methylesterase in complex with hexasaccharide V | | Descriptor: | Pectinesterase A, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-07 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
2NZM
 
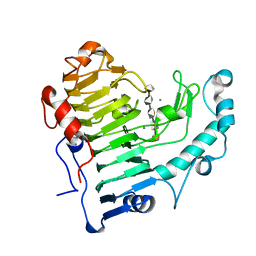 | | Hexasaccharide I bound to Bacillus subtilis pectate lyase | | Descriptor: | CALCIUM ION, Pectate lyase, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Pickersgill, R.W, Seyedarabi, A. | | Deposit date: | 2006-11-24 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into substrate specificity and the anti beta-elimination mechanism of pectate lyase.
Biochemistry, 49, 2010
|
|
2O04
 
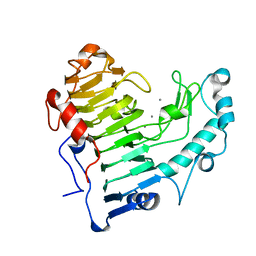 | | Pectate lyase bound to hexasaccharide compound II | | Descriptor: | CALCIUM ION, Pectate lyase, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Pickersgill, R.W, Seyedarabi, A. | | Deposit date: | 2006-11-27 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into substrate specificity and the anti beta-elimination mechanism of pectate lyase.
Biochemistry, 49, 2010
|
|
2MCV
 
 | | Solid-state NMR structure of piscidin 1 in aligned 1:1 phosphatidylethanolamine/phosphoglycerol lipid bilayers | | Descriptor: | Moronecidin | | Authors: | Fu, R, Tian, Y, Perrin Jr, B.S, Grant, C.V, Hayden, R.M, Pastor, R.W, Cotten, M.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-19 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution structures and orientations of antimicrobial peptides piscidin 1 and piscidin 3 in fluid bilayers reveal tilting, kinking, and bilayer immersion.
J.Am.Chem.Soc., 136, 2014
|
|
1K6A
 
 | | Structural studies on the mobility in the active site of the Thermoascus aurantiacus xylanase I | | Descriptor: | xylanase I | | Authors: | Lo Leggio, L, Kalogiannis, S, Eckert, K, Teixeira, S.C.M, Bhat, M.K, Andrei, C, Pickersgill, R.W, Larsen, S. | | Deposit date: | 2001-10-15 | | Release date: | 2002-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Substrate specificity and subsite mobility in T. aurantiacus xylanase 10A.
FEBS LETT., 509, 2001
|
|
2NT6
 
 | | Crystal structure of pectin methylesterase D178A mutant in complex with hexasaccharide III | | Descriptor: | Pectinesterase A, methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-methyl alpha-D-galactopyranuronate | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-07 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
2O1D
 
 | | Pectate lyase bound to trisaccharide | | Descriptor: | CALCIUM ION, Pectate lyase, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Pickersgill, R.W, Seyedarabi, A. | | Deposit date: | 2006-11-28 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into substrate specificity and the anti beta-elimination mechanism of pectate lyase.
Biochemistry, 49, 2010
|
|
2K5X
 
 | |
2O0V
 
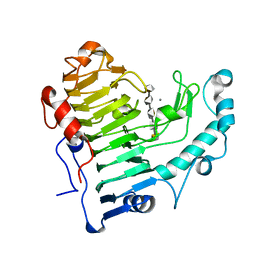 | | Pectate lyase bound to hexasaccharide compound III | | Descriptor: | CALCIUM ION, Pectate lyase, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Pickersgill, R.W, Seyedarabi, A. | | Deposit date: | 2006-11-28 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into substrate specificity and the anti beta-elimination mechanism of pectate lyase.
Biochemistry, 49, 2010
|
|
2N06
 
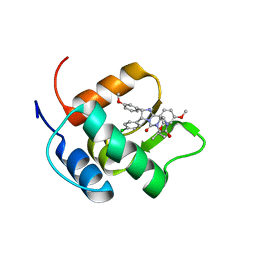 | | Mdmx-298 | | Descriptor: | 4-[[(4S,5R)-5-(4-chlorophenyl)-4-(3-methoxyphenyl)-2-(4-methoxy-2-propan-2-yloxy-phenyl)-4,5-dihydroimidazol-1-yl]carbonyl]piperazin-2-one, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
1DUE
 
 | |
1DT2
 
 | |
1DUA
 
 | |
1EZJ
 
 | | CRYSTAL STRUCTURE OF THE MULTIMERIZATION DOMAIN OF THE PHOSPHOPROTEIN FROM SENDAI VIRUS | | Descriptor: | CALCIUM ION, ETHYL MERCURY ION, NUCLEOCAPSID PHOSPHOPROTEIN | | Authors: | Tarbouriech, N, Curran, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2000-05-11 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric coiled coil domain of Sendai virus phosphoprotein.
Nat.Struct.Biol., 7, 2000
|
|
