2CAL
 
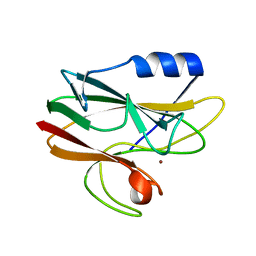 | | Crystal structure of His143Met rusticyanin | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Barrett, M.L, Harvey, I, Sundararajan, M, Surendran, R, Hall, J.F, Ellis, M.J, Hough, M.A, Strange, R.W, Hillier, I.H, Hasnain, S.S. | | Deposit date: | 2005-12-21 | | Release date: | 2006-01-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Crystal Structures, Exafs, and Quantum Chemical Studies of Rusticyanin and its Two Mutants Provide Insight Into its Unusual Properties.
Biochemistry, 45, 2006
|
|
2C9S
 
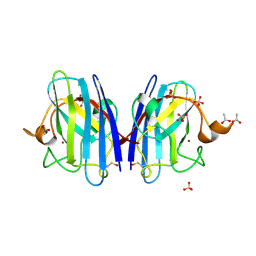 | | 1.24 Angstroms resolution structure of Zn-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2C9G
 
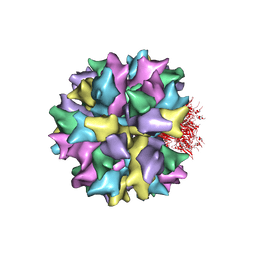 | | THE QUASI-ATOMIC MODEL OF THE ADENOVIRUS TYPE 3 PENTON BASE DODECAHEDRON | | Descriptor: | PENTON PROTEIN | | Authors: | Fuschiotti, P, Schoehn, G, Fender, P, Fabry, C.M.S, Hewat, E.A, Chroboczek, J, Ruigrok, R.W.H, Conway, J.F. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Structure of the Dodecahedral Penton Particle from Human Adenovirus Type 3.
J.Mol.Biol., 356, 2006
|
|
2CAK
 
 | | 1.27Angstrom Structure of Rusticyanin from Thiobacillus ferrooxidans | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Barrett, M.L, Harvey, I, Sundararajan, M, Surendran, R, Hall, J.F, Ellis, M.J, Hough, M.A, Strange, R.W, Hillier, I.H, Hasnain, S.S. | | Deposit date: | 2005-12-21 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Atomic Resolution Crystal Structures, Exafs, and Quantum Chemical Studies of Rusticyanin and its Two Mutants Provide Insight Into its Unusual Properties.
Biochemistry, 45, 2006
|
|
2B6A
 
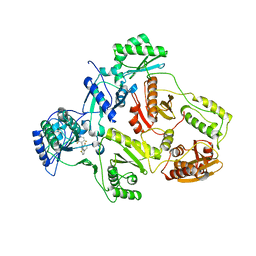 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with THR-50 | | Descriptor: | 1-(2,6-DIFLUOROBENZYL)-2-(2,6-DIFLUOROPHENYL)-4-METHYL-1H-BENZIMIDAZOLE, Reverse transcriptase p51 subunit, Reverse transcriptase p66 subunit | | Authors: | Morningstar, M.L, Roth, T, Smith, M.K, Zajac, M, Watson, K, Buckheit, R.W, Das, K, Zhang, W, Arnold, E, Michejda, C.J. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with THR-50
TO BE PUBLISHED
|
|
2AOI
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P1-P6 | | Descriptor: | PEPTIDE INHIBITOR, POL POLYPROTEIN, SULFATE ION | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2AVM
 
 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, AND G73S | | Descriptor: | ACETIC ACID, GLYCEROL, HIV-1 protease, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2AVS
 
 | | kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, and G73S | | Descriptor: | ACETIC ACID, DIMETHYL SULFOXIDE, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2AOH
 
 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P6-PR | | Descriptor: | CHLORIDE ION, PEPTIDE INHIBITOR, POL POLYPROTEIN, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2B5T
 
 | | 2.1 Angstrom structure of a nonproductive complex between antithrombin, synthetic heparin mimetic SR123781 and two S195A thrombin molecules | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfonato-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-alpha-L-idopyranuronic acid-(1-4)-methyl 3-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Johnson, D.J, Li, W, Luis, S.A, Carrell, R.W, Huntington, J.A. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation.
J.Biol.Chem., 281, 2006
|
|
1ZKY
 
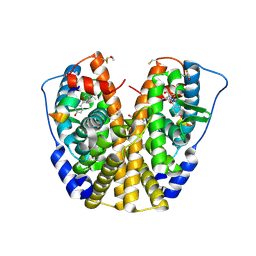 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-3M and A Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide | | Descriptor: | 4-[(1S,2S,5S)-5-(HYDROXYMETHYL)-6,8,9-TRIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Hahm, J.B, Nettles, K.W, Greene, G.L. | | Deposit date: | 2005-05-04 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity.
J.Biol.Chem., 281, 2006
|
|
2AOE
 
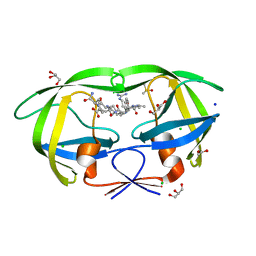 | | crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog CA-P2 | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2ANT
 
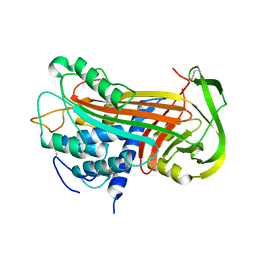 | | THE 2.6 A STRUCTURE OF ANTITHROMBIN INDICATES A CONFORMATIONAL CHANGE AT THE HEPARIN BINDING SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose, ANTITHROMBIN | | Authors: | Skinner, R, Abrahams, J.-P, Whisstock, J.C, Lesk, A.M, Carrell, R.W, Wardell, M.R. | | Deposit date: | 1997-01-28 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6 A structure of antithrombin indicates a conformational change at the heparin binding site.
J.Mol.Biol., 266, 1997
|
|
2AVV
 
 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, and G73S | | Descriptor: | ACETIC ACID, CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2AOF
 
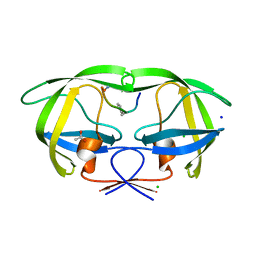 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P1-P6 | | Descriptor: | ACETIC ACID, CHLORIDE ION, PEPTIDE INHIBITOR, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2B23
 
 | | Human estrogen receptor alpha ligand-binding domain and a glucocorticoid receptor-interacting protein 1 NR box II peptide | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
2C9U
 
 | | 1.24 Angstroms resolution structure of as-isolated Cu-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2CEO
 
 | | thyroxine-binding globulin complex with thyroxine | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, GLYCEROL, THYROXINE-BINDING GLOBULIN | | Authors: | Zhou, A, Wei, Z, Read, R.J, Carrell, R.W. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Mechanism for the Carriage and Release of Thyroxine in the Blood.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2DCO
 
 | |
1GC1
 
 | | HIV-1 GP120 CORE COMPLEXED WITH CD4 AND A NEUTRALIZING HUMAN ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, CD4, ... | | Authors: | Kwong, P.D, Wyatt, R, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 1998-06-15 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody.
Nature, 393, 1998
|
|
1FWS
 
 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP AND CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-24 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
1FXP
 
 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, PHOSPHATE ION | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-26 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate and metal complexes of 3-deoxy-D-manno-octulosonate-8-phosphate synthase from Aquifex aeolicus at 1.9-A resolution. Implications for the condensation mechanism.
J.Biol.Chem., 276, 2001
|
|
1HN3
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL 37 AMINO ACIDS OF THE MOUSE ARF TUMOR SUPPRESSOR PROTEIN | | Descriptor: | P19 ARF PROTEIN | | Authors: | DiGiammarino, E.L, Filippov, I, Weber, J.D, Bothner, B, Kriwacki, R.W. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the p53 regulatory domain of the p19Arf tumor suppressor protein.
Biochemistry, 40, 2001
|
|
1HYO
 
 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH 4-(HYDROXYMETHYLPHOSPHINOYL)-3-OXO-BUTANOIC ACID | | Descriptor: | 4-[HYDROXY-[METHYL-PHOSPHINOYL]]-3-OXO-BUTANOIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Bateman, R.L, Bhanumoorthy, P, Witte, J.F, McClard, R.W, Grompe, M, Timm, D.E. | | Deposit date: | 2001-01-21 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanistic inferences from the crystal structure of fumarylacetoacetate hydrolase with a bound phosphorus-based inhibitor.
J.Biol.Chem., 276, 2001
|
|
1I1S
 
 | | SOLUTION STRUCTURE OF THE TRANSCRIPTIONAL ACTIVATION DOMAIN OF THE BACTERIOPHAGE T4 PROTEIN MOTA | | Descriptor: | MOTA | | Authors: | Li, N, Zhang, W, White, S.W, Kriwacki, R.W. | | Deposit date: | 2001-02-02 | | Release date: | 2001-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transcriptional activation domain of the bacteriophage T4 protein, MotA.
Biochemistry, 40, 2001
|
|
