6YLT
 
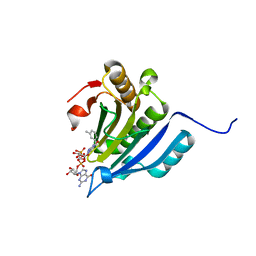 | | Translation initiation factor 4E in complex with 3-MeBn7GpppG mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, [[(2~{R},3~{S},4~{R},5~{R})-5-[2-azanyl-7-[(3-methylphenyl)methyl]-6-oxidanylidene-1~{H}-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] hydrogen phosphate | | Authors: | Kubacka, D, Wojcik, R, Baranowski, M.R, Kowalska, J, Jemielity, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Novel N7-Arylmethyl Substituted Dinucleotide mRNA 5' cap Analogs: Synthesis and Evaluation as Modulators of Translation.
Pharmaceutics, 13, 2021
|
|
6OXY
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-19 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
6P11
 
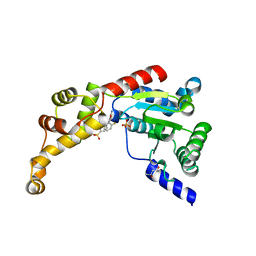 | | Structure of spastin AAA domain (T692A mutant) in complex with JNJ-7706621 inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Drosophila melanogaster Spastin AAA domain, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Analyzing Resistance to Design Selective Chemical Inhibitors for AAA Proteins.
Cell Chem Biol, 26, 2019
|
|
8GOD
 
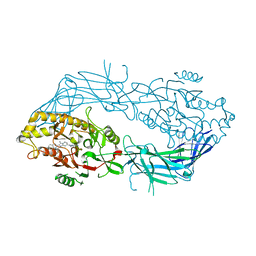 | | Co-crystal structure of Human Protein-arginine deiminase type-4 (PAD4) with small molecule inhibitor JBI-589 | | Descriptor: | Protein-arginine deiminase type-4, [(3~{R})-3-azanylpiperidin-1-yl]-[2-[1-[(4-fluorophenyl)methyl]indol-2-yl]-3-methyl-imidazo[1,2-a]pyridin-7-yl]methanone | | Authors: | Swaminathan, S, Birudukota, S, Vaithilingam, K, Kandan, S, Asaithambi, K, Kathiresan, N, Gosu, R, Rajagopal, S, Sadhu, N. | | Deposit date: | 2022-08-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Alleviation of arthritis through prevention of neutrophil extracellular traps by an orally available inhibitor of protein arginine deiminase 4.
Sci Rep, 13, 2023
|
|
1UVT
 
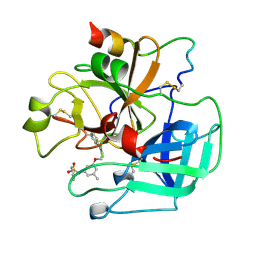 | | BOVINE THROMBIN--BM14.1248 COMPLEX | | Descriptor: | N-{3-METHYL-5-[2-(PYRIDIN-4-YLAMINO)-ETHOXY]-PHENYL}-BENZENESULFONAMIDE, THROMBIN | | Authors: | Engh, R.A, Huber, R. | | Deposit date: | 1996-10-16 | | Release date: | 1997-11-19 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme flexibility, solvent and 'weak' interactions characterize thrombin-ligand interactions: implications for drug design.
Structure, 4, 1996
|
|
6P5J
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 2) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
1UVS
 
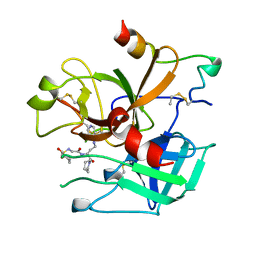 | | BOVINE THROMBIN--BM51.1011 COMPLEX | | Descriptor: | THROMBIN, [[CYCLOHEXANESULFONYL-GLYCYL]-3[PYRIDIN-4-YL-AMINOMETHYL]ALANYL]PIPERIDINE | | Authors: | Engh, R.A, Huber, R. | | Deposit date: | 1996-10-16 | | Release date: | 1997-11-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme flexibility, solvent and 'weak' interactions characterize thrombin-ligand interactions: implications for drug design.
Structure, 4, 1996
|
|
1K6F
 
 | |
7TOK
 
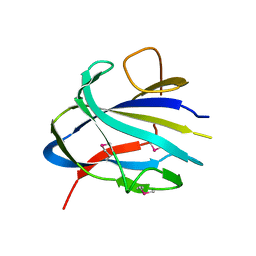 | | Crystal structure of the CBM domain of carbohydrate esterase FjoAcXE | | Descriptor: | Acetylxylan esterase I | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
1UVU
 
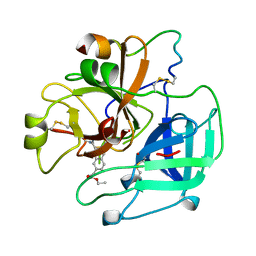 | | BOVINE THROMBIN--BM12.1700 COMPLEX | | Descriptor: | 3-(7-DIAMINOMETHYL-NAPHTHALEN-2-YL)-PROPIONIC ACID ETHYL ESTER, THROMBIN | | Authors: | Engh, R.A, Huber, R. | | Deposit date: | 1996-10-16 | | Release date: | 1997-11-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme flexibility, solvent and 'weak' interactions characterize thrombin-ligand interactions: implications for drug design.
Structure, 4, 1996
|
|
8H7E
 
 | | Crystal structure of a de novo enzyme, ferric enterobactin esterase Syn-F4 (K4T) at 2.0 angstrom resolution | | Descriptor: | ACETATE ION, De novo ferric enterobactin esterase Syn-F4 | | Authors: | Kurihara, K, Umezawa, K, Donnelly, A.E, Hecht, M.H, Arai, R. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and activity of a de novo enzyme, ferric enterobactin esterase Syn-F4.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D93
 
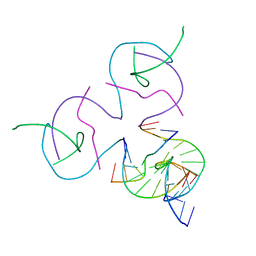 | | [2T7] Self-assembling tensegrity triangle with R3 symmetry at 2.96 A resolution, update and junction cut for entry 3GBI | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*TP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*AP*CP*AP*CP*CP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Woloszyn, K, Lu, B, Sha, R, Ohayon, Y.P, Seeman, N.C. | | Deposit date: | 2022-06-09 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The Rule of Thirds: Controlling Junction Chirality and Polarity in 3D DNA Tiles.
Small, 19, 2023
|
|
7UL8
 
 | | [T:Au+:T--pH 11] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2022-04-04 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4.22 Å) | | Cite: | Metal-Mediated DNA Nanotechnology in 3D: Structural Library by Templated Diffraction.
Adv Mater, 35, 2023
|
|
8HUA
 
 | | Serial synchrotron crystallography structure of ba3-type cytochrome c oxidase from Thermus thermophilus using a goniometer compatible flow-cell | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Ghosh, S, Zoric, D, Bjelcic, M, Johannesson, J, Sandelin, E, Branden, G, Neutze, R. | | Deposit date: | 2022-12-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A simple goniometer-compatible flow cell for serial synchrotron X-ray crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
6IDG
 
 | | antibody 64M-5 Fab in complex with dT(6-4)T | | Descriptor: | Anti-(6-4) photoproduct antibody 64M-5 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-5 Fab (light chain), DNA (5'-D(*(64T)P*(5PY))-3') | | Authors: | Yokoyama, H, Mizutani, R, Noguchi, S, Hayashida, N. | | Deposit date: | 2018-09-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the antibody 64M-5 Fab and its complex with dT(6-4)T indicate induced-fit and high-affinity mechanisms.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8EQJ
 
 | | Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like membrane environment, MSP1D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8EQU
 
 | | Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like environment, Saposin A nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein, Saposin A, ... | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8I5A
 
 | | N-acetyl-(R)-beta-phenylalanine acylase, 2.75 angstrom resolution | | Descriptor: | N-acetyl-(R)-beta-phenylalanine acylase | | Authors: | Kato, Y, Natsume, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Expression, purification and crystallization of N-acetyl-(R)-beta-phenylalanine acylases derived from Burkholderia sp. AJ110349 and Variovorax sp. AJ110348 and structure determination of the Burkholderia enzyme.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8EQT
 
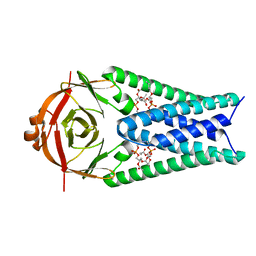 | | Structure of SARS-CoV-2 Orf3a in plasma membrane-like environment, MSP1D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8I59
 
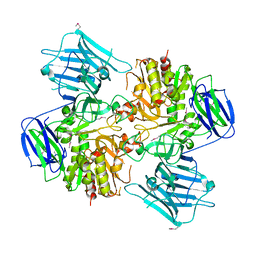 | | N-acetyl-(R)-beta-phenylalanine acylase, selenomethionyl derivative | | Descriptor: | N-acetyl-(R)-beta-phenylalanine acylase | | Authors: | Kato, Y, Natsume, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Expression, purification and crystallization of N-acetyl-(R)-beta-phenylalanine acylases derived from Burkholderia sp. AJ110349 and Variovorax sp. AJ110348 and structure determination of the Burkholderia enzyme.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8HUY
 
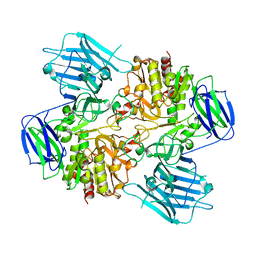 | | N-acetyl-(R)-beta-phenylalanine acylase | | Descriptor: | Chains: A,B | | Authors: | Kato, Y, Natsume, R. | | Deposit date: | 2022-12-24 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expression, purification and crystallization of N-acetyl-(R)-beta-phenylalanine acylases derived from Burkholderia sp. AJ110349 and Variovorax sp. AJ110348 and structure determination of the Burkholderia enzyme.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8EQS
 
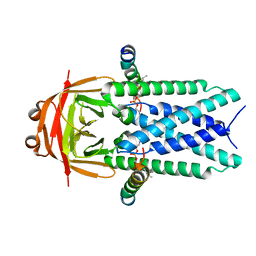 | | Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8I43
 
 | |
8I44
 
 | |
