6X1S
 
 | | Structure of pHis Fab (SC1-1) in complex with pHis mimetic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NM23-1-pTza peptide, SC1-1 Heavy chain, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Wilson, I.A, Hunter, T. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for differential recognition of phosphohistidine-containing peptides by 1-pHis and 3-pHis monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6NYV
 
 | | Structure of spastin AAA domain in complex with a quinazoline-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, N-(3-tert-butyl-1H-pyrazol-5-yl)-2-[(2R)-2-methylpiperazin-1-yl]quinazolin-4-amine, SULFATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | Designing Allele-Specific Inhibitors of Spastin, a Microtubule-Severing AAA Protein.
J. Am. Chem. Soc., 141, 2019
|
|
8GRT
 
 | | Small Dipeptide Analogues developed by Co-crystal Structure of Stenotrophomonas maltophilia Dipeptidyl Peptidase 7 | | Descriptor: | 2-AMINO-3-CYCLOHEXYL-PROPIONIC ACID, Dipeptidyl-peptidase, TYROSINE | | Authors: | Yasumitsu, S, Koushi, H, Akihiro, N, Yoshiyuki, Y, Wataru, O, Mizuki, S, Saori, R, Nobutada, T, Anna, M, Keiko, H, Tsuda, Y. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Small Dipeptide Analogues Generated by Co-crystal Structure of Bacterial Dipeptidyl Peptidase 7 to Defeat Stenotrophomonas maltophilia
To Be Published
|
|
4J2I
 
 | | Multiple crystal structures of an all-AT DNA dodecamer stabilized by weak interactions | | Descriptor: | 5'-D(*AP*AP*TP*AP*AP*AP*TP*TP*TP*AP*TP*T)-3' | | Authors: | Acosta-Reyes, F.J, Subirana, J.A, Pous, J, Sanchez, R, Condom, N, Baldini, R, Malinina, L, Campos, J.L. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Polymorphic crystal structures of an all-AT DNA dodecamer.
Biopolymers, 103, 2015
|
|
6O0X
 
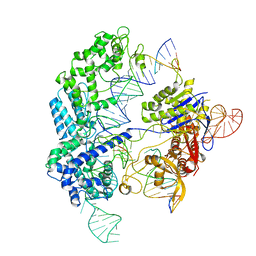 | | Conformational states of Cas9-sgRNA-DNA ternary complex in the presence of magnesium | | Descriptor: | 3' product of target strand DNA, 5' product of target strand DNA, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Zhu, X, Clarke, R, Puppala, A.K, Chittori, S, Merk, A, Merrill, B.J, Simonovic, M, Subramaniam, S. | | Deposit date: | 2019-02-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6O26
 
 | | Crystal structure of 3246 Fab in complex with circumsporozoite protein NANA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3246 Fab heavy chain, ... | | Authors: | Scally, S.W, Bosch, A, Castro, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-03-04 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
6WLE
 
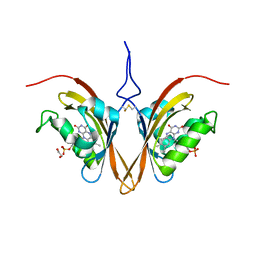 | | Crystal structure of the Zeitlupe light-state mimic G46A | | Descriptor: | 1,2-ETHANEDIOL, Adagio protein 1, FLAVIN MONONUCLEOTIDE | | Authors: | Zoltowski, B, Green, R. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Steric and Electronic Interactions at Gln154 in ZEITLUPE Induce Reorganization of the LOV Domain Dimer Interface.
Biochemistry, 60, 2021
|
|
6WLP
 
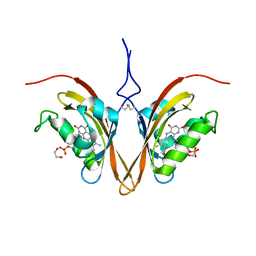 | | Crystal Structure of the ZTL light-state mimic G46S | | Descriptor: | 1,2-ETHANEDIOL, Adagio protein 1, FLAVIN MONONUCLEOTIDE | | Authors: | Zoltowski, B, Green, R. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Steric and Electronic Interactions at Gln154 in ZEITLUPE Induce Reorganization of the LOV Domain Dimer Interface.
Biochemistry, 60, 2021
|
|
8GLH
 
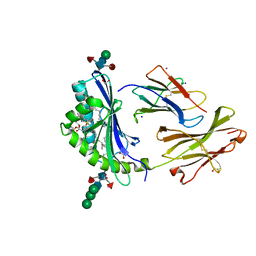 | | Crystal Structure of Human CD1b in Complex with Endogenous PC C40:5 | | Descriptor: | (11E)-hexadec-11-enoic acid, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Farquhar, R, Rossjohn, J, Shahine, A. | | Deposit date: | 2023-03-22 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms.
Cell, 186, 2023
|
|
6NRA
 
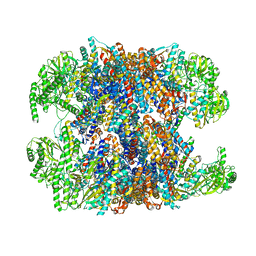 | | hTRiC-hPFD Class1 (No PFD) | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
8G6B
 
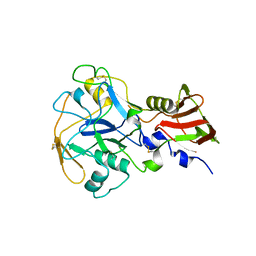 | | Crystal structure of PfAMA1-RON2L chimera | | Descriptor: | Apical membrane antigen 1, rhoptry neck protein 2 chimera, SULFATE ION | | Authors: | Boulanger, M.J, Ramaswamy, R. | | Deposit date: | 2023-02-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure guided mimicry of an essential P. falciparum receptor-ligand complex enhances cross neutralizing antibodies.
Nat Commun, 14, 2023
|
|
6WCG
 
 | |
6WCN
 
 | |
6WCL
 
 | |
6WCP
 
 | |
6WCM
 
 | |
6NSI
 
 | | Crystal structure of Fe(III)-bound YtgA from Chlamydia trachomatis | | Descriptor: | CALCIUM ION, FE (III) ION, Manganese-binding protein, ... | | Authors: | Luo, Z, Campbell, R, Begg, S.L, Kobe, B, McDevitt, C.A. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.00006342 Å) | | Cite: | Structure and Metal Binding Properties of Chlamydia trachomatis YtgA.
J.Bacteriol., 202, 2019
|
|
6WCH
 
 | |
6O2C
 
 | | Crystal structure of 4493 Fab in complex with circumsporozoite protein NANP3 and anti-kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, 4493 Fab heavy chain, 4493 Kappa light chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
6NBS
 
 | | WT ERK2 with compound 2507-8 | | Descriptor: | (5S)-5-benzyl-4,5-dihydro-1H-imidazol-2-amine, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sammons, R.M, Perry, N.A, Cho, E.J, Kaoud, T.S, Zamora-Olivares, D.P, Piserchio, A, Houghten, R.A, Giulianotti, M, Li, Y, Debevec, G, Gurevich, V.V, Ghose, R, Iverson, T.M, Dalby, K.N. | | Deposit date: | 2018-12-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Class of Common Docking Domain Inhibitors That Prevent ERK2 Activation and Substrate Phosphorylation.
Acs Chem.Biol., 14, 2019
|
|
6NX4
 
 | | Structure of the C-terminal Helical Repeat Domain of Eukaryotic Elongation Factor 2 Kinase (eEF-2K) | | Descriptor: | Eukaryotic elongation factor 2 kinase | | Authors: | Piserchio, A, Will, N, Giles, D.H, Hajredini, F, Dalby, K.N, Ghose, R. | | Deposit date: | 2019-02-08 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Carboxy-Terminal Tandem Repeat Domain of Eukaryotic Elongation Factor 2 Kinase and Its Role in Substrate Recognition.
J.Mol.Biol., 431, 2019
|
|
6WH6
 
 | | Crystal structure of human sulfide quinone oxidoreductase in complex with coenzyme Q (cyanide soaked) | | Descriptor: | CYANIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Sulfide:quinone oxidoreductase, ... | | Authors: | Banerjee, R, Cho, U.S, Moon, S. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dismantling and Rebuilding the Trisulfide Cofactor Demonstrates Its Essential Role in Human Sulfide Quinone Oxidoreductase.
J.Am.Chem.Soc., 142, 2020
|
|
7UQ3
 
 | | JmjC domain-containing protein 5 (JMJD5) in complex with Mn and (S)-2-(1-hydroxy-2,5-dioxopyrrolidin-3-yl)acetic acid | | Descriptor: | Bifunctional peptidase and arginyl-hydroxylase JMJD5, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Chowdhury, R, Islam, M.S, Schofield, C.J. | | Deposit date: | 2022-04-19 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural analysis of the 2-oxoglutarate binding site of the circadian rhythm linked oxygenase JMJD5.
Sci Rep, 12, 2022
|
|
8G5V
 
 | | Empty capsid of Hepatitis B virus | | Descriptor: | Core protein Cp183 | | Authors: | Yang, R, Cingolani, G. | | Deposit date: | 2023-02-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for nuclear import of hepatitis B virus (HBV) nucleocapsid core.
Sci Adv, 10, 2024
|
|
8G6V
 
 | |
