8QWR
 
 | | Crystal structure of CotB2 variant V80L | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Cyclooctat-9-en-7-ol synthase, ... | | Authors: | Dimos, N, Himpich, S, Ringel, M, Driller, R, Major, D.T, Brueck, T, Loll, B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of CotB2 variant V80L
To Be Published
|
|
8QWS
 
 | | Crystal structure of CotB2 variant V80L in complex with alendronate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Dimos, N, Himpich, S, Ringel, M, Driller, R, Major, D.T, Brueck, T, Loll, B. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CotB2 variant V80L in complex with alendronate
To Be Published
|
|
5JMZ
 
 | |
5JN6
 
 | |
5J17
 
 | |
8QUU
 
 | | Crystal structure of chlorite dismutase at 3000 eV based on spherical harmonics absorption corrections | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Duman, R, Wagner, A, Kamps, J, Orville, A. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QVV
 
 | | Crystal structure of Ompk36 GD at 3500 eV based on analytical absorption corrections | | Descriptor: | OmpK36, SULFATE ION | | Authors: | Duman, R, Wagner, A, Beis, K, Wong, J. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
5JPZ
 
 | | Crystal structure of HAT domain of human Squamous Cell Carcinoma Antigen Recognized By T Cells 3, SART3 (TIP110) | | Descriptor: | Squamous cell carcinoma antigen recognized by T-cells 3 | | Authors: | Grazette, A, Harper, S, Emsley, J, Layfield, R, Dreveny, I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.045 Å) | | Cite: | unpublished
To Be Published
|
|
2MTL
 
 | | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109 | | Descriptor: | De novo designed protein FR55 OR109 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Ciccosanti, C, Sahdev, S, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109
To be Published
|
|
8QAA
 
 | |
8QVB
 
 | | Crystal structure of chlorite dismutase at 3000 eV based on a combination of spherical harmonics and analytical absorption corrections | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Duman, R, Wagner, A, Kamps, J, Orville, A. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QUQ
 
 | | Crystal structure of Ompk36 GD at 3500 eV based on spherical harmonics absorption corrections | | Descriptor: | OmpK36, SULFATE ION | | Authors: | Duman, R, Wagner, A, Beis, K, Wong, J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
5JTE
 
 | | Cryo-EM structure of an ErmBL-stalled ribosome in complex with A-, P-, and E-tRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Arenz, S, Bock, L.V, Graf, M, Innis, C.A, Beckmann, R, Grubmueller, H, Vaiana, A.C, Wilson, D.N. | | Deposit date: | 2016-05-09 | | Release date: | 2016-07-20 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest.
Nat Commun, 7, 2016
|
|
8QE4
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*CP*(FRG)P*CP*(FRG)P*CP*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QUR
 
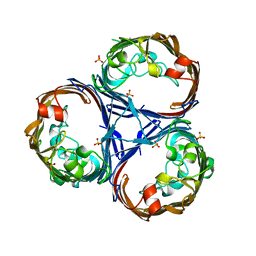 | | Crystal structure of Ompk36 GD at 3500 eV with no absorption corrections | | Descriptor: | OmpK36, SULFATE ION | | Authors: | Duman, R, Wagner, A, Beis, K, Wong, J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
8QOR
 
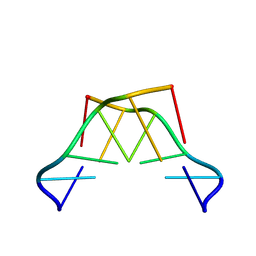 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(CFL)P*(FRG)P*(CFL)P*(FRG)P*(CFL)P*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QAI
 
 | |
8QAH
 
 | |
8QKD
 
 | |
8QDU
 
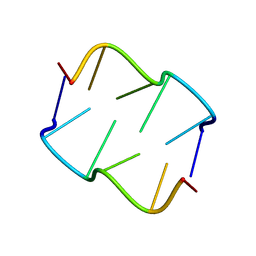 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(FC)P*GP*(FC)P*GP*(FC)P*G)-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M.J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QAB
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil hexameric alpha-helical barrel with 4 heptad repeats, apCCHex | | Descriptor: | 1-methoxy-2-[2-[2-[2-[2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethane, apCCHex | | Authors: | Naudin, E.N, Albanese, K.I, Petrenas, R, Woolfson, D.N. | | Deposit date: | 2023-08-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rationally seeded computational protein design of ɑ-helical barrels.
Nat.Chem.Biol., 20, 2024
|
|
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5JP4
 
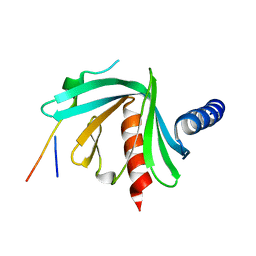 | |
8QAD
 
 | |
8QUV
 
 | | Crystal structure of chlorite dismutase at 3000 eV with no absorption corrections | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Duman, R, Wagner, A, Kamps, J, Orville, A. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
