1T6S
 
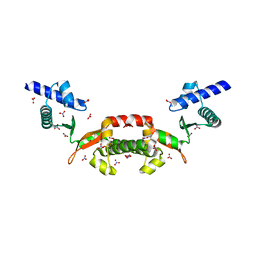 | | Crystal structure of a conserved hypothetical protein from Chlorobium tepidum | | Descriptor: | NITRATE ION, conserved hypothetical protein | | Authors: | Kim, J.S, Shin, D.H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-05-07 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of ScpB from Chlorobium tepidum, a protein involved in chromosome partitioning.
Proteins, 62, 2006
|
|
6XEA
 
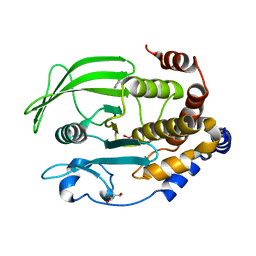 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 3 bound to vanadate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
1T7C
 
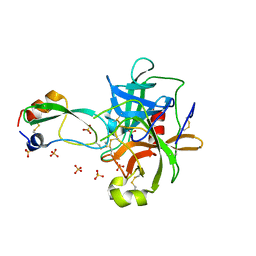 | | CRYSTAL STRUCTURE OF THE P1 GLU BPTI MUTANT- BOVINE CHYMOTRYPSIN COMPLEX | | Descriptor: | Chymotrypsin A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Czapinska, H, Helland, R, Otlewski, J, Smalas, A.O. | | Deposit date: | 2004-05-09 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of five bovine chymotrypsin complexes with P1 BPTI variants.
J.Mol.Biol., 344, 2004
|
|
6XEE
 
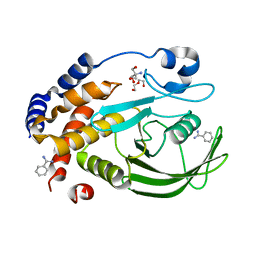 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 4 apo form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
8HRG
 
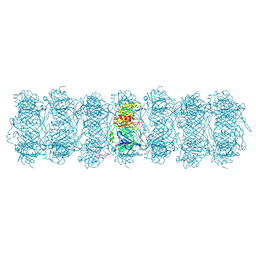 | | Tail tube of DT57C bacteriophage in the full state | | Descriptor: | Tail tube protein | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
6XE8
 
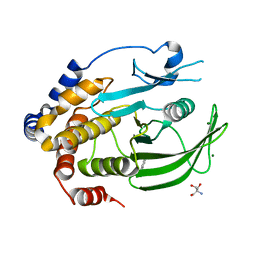 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 3 apo form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XED
 
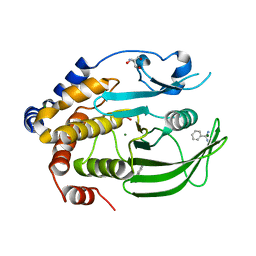 | | Crystal Structure of the PTP1B YopH WPD loop Chimera 3 bound to tungstate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
6XEF
 
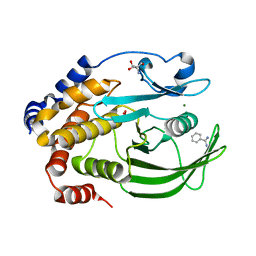 | | Crystal structure of the PTP1B YopH WPD loop Chimera 4 bound to vanadate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.048 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
4YYP
 
 | | Crystal structure of human PLK4-PB3 in complex with STIL-CC | | Descriptor: | CHLORIDE ION, SCL-interrupting locus protein, Serine/threonine-protein kinase PLK4 | | Authors: | Imseng, S, Arquint, C, Gabryjonczyk, A, Boehm, R, Sauer, E, Hiller, S, Nigg, E.A, Maier, T. | | Deposit date: | 2015-03-24 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | STIL binding to Polo-box 3 of PLK4 regulates centriole duplication.
Elife, 4, 2015
|
|
6XEG
 
 | | Crystal structure of the PTP1B YopH WPD loop Chimera 4 bound to tungstate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, MAGNESIUM ION, ... | | Authors: | Olsen, K.J, Shen, R, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Insights into the importance of WPD-loop sequence for activity and structure in protein tyrosine phosphatases.
Chem Sci, 13, 2022
|
|
5DAT
 
 | | Complex of yeast 80S ribosome with hypusine-containing eIF5A | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
8HRE
 
 | | Straight fiber of DT57C bacteriophage in the full state | | Descriptor: | Central straight fiber | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8F41
 
 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
8HQK
 
 | | Capsid of DT57C bacteriophage in the empty state | | Descriptor: | Major head protein | | Authors: | Ayala, R, Moiseenko, A.V, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8HOY
 
 | | Cryo-EM structure of monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex without DNA at 2.76 angostram | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures of human monkeypox viral replication complexes with and without DNA duplex.
Cell Res., 33, 2023
|
|
6OQ8
 
 | |
8HO3
 
 | | Capsid of DT57C bacteriophage in the full state | | Descriptor: | Major head protein | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-09 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8HQZ
 
 | | Baseplate of DT57C bacteriophage in the full state | | Descriptor: | Baseplate hub protein, Distal tail protein, L-shaped tail fiber assembly, ... | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-14 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
8HQO
 
 | | Neck of DT57C bacteriophage in the full state | | Descriptor: | Head completion protein, Portal protein, Tail terminator protein, ... | | Authors: | Ayala, R, Moiseenko, A.V, Chen, T.H, Kulikov, E.E, Golomidova, A.K, Orekhov, P.S, Street, M.A, Sokolova, O.S, Letarov, A.V, Wolf, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers.
Nat Commun, 14, 2023
|
|
6OUM
 
 | |
6Y94
 
 | | Ca2+-bound Calmodulin mutant N53I | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Holt, C, Nielsen, L.H, Lau, K, Brohus, M, Sorensen, A.B, Larsen, K.T, Sommer, C, Petegem, F.V, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2020-03-06 | | Release date: | 2020-04-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The arrhythmogenic N53I variant subtly changes the structure and dynamics in the calmodulin N-terminal domain, altering its interaction with the cardiac ryanodine receptor.
J.Biol.Chem., 295, 2020
|
|
6OJH
 
 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 7-[(3R)-3-aminopyrrolidin-1-yl]-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine, ACETATE ION, Biotin carboxylase, ... | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine
To Be Published
|
|
6OEZ
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-N-(Cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrro-lidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, N-(cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
6OR0
 
 | | Crystal structure of Insulin from Non-merohedrally twinned crystals | | Descriptor: | Insulin chain A, Insulin chain B | | Authors: | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | Deposit date: | 2019-04-29 | | Release date: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OUD
 
 | |
