6G0B
 
 | |
2VGU
 
 | | Crystal structure of E53QbsSHMT with L-serine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Rajaram, V, Bhavani, B.S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2007-11-15 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure Determination and Biochemical Studies on Bacillus Stearothermophilus E53Q Serine Hydroxymethyltransferase and its Complexes Provide Insights on Function and Enzyme Memory
FEBS J., 274, 2007
|
|
5NGF
 
 | | Crystal structure of USP7 in complex with the covalent inhibitor, FT827 | | Descriptor: | 1,2-ETHANEDIOL, Ubiquitin carboxyl-terminal hydrolase 7, ~{N}-[2-[4-[4-[(1-methyl-4-oxidanylidene-pyrazolo[3,4-d]pyrimidin-5-yl)methyl]-4-oxidanyl-piperidin-1-yl]carbonylphenyl]phenyl]ethanesulfonamide | | Authors: | Krajewski, W.W, Turnbull, A.P, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
5IOO
 
 | | Accommodation of massive sequence variation in Nanoarchaeota by the C-type lectin fold | | Descriptor: | AvpA, MAGNESIUM ION | | Authors: | Handa, S, Paul, B, Miller, J, Valentine, D, Ghosh, P. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Conservation of the C-type lectin fold for accommodating massive sequence variation in archaeal diversity-generating retroelements.
Bmc Struct.Biol., 16, 2016
|
|
5ISC
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0491 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0491
To Be Published
|
|
5NE1
 
 | | L2 class A serine-beta-lactamase in complex with cyclic boronate 2 | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, Beta-lactamase, TRIETHYLENE GLYCOL | | Authors: | Hinchliffe, P, Calvopina, K, Spencer, J. | | Deposit date: | 2017-03-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural/mechanistic insights into the efficacy of nonclassical beta-lactamase inhibitors against extensively drug resistant Stenotrophomonas maltophilia clinical isolates.
Mol. Microbiol., 106, 2017
|
|
2VL8
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF LETHAL TOXIN FROM CLOSTRIDIUM SORDELLII IN COMPLEX WITH UDP, CASTANOSPERMINE AND CALCIUM ION | | Descriptor: | CALCIUM ION, CASTANOSPERMINE, CYTOTOXIN L, ... | | Authors: | Jank, T, Ziegler, M.O.P, Schulz, G.E, Aktories, K. | | Deposit date: | 2008-01-09 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Inhibition of the Glucosyltransferase Activity of Clostridial Rho/Ras-Glucosylating Toxins by Castanospermine.
FEBS Lett., 582, 2008
|
|
7AZK
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 35 bound | | Descriptor: | Beta sliding clamp, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
4AOM
 
 | | MTIP and MyoA complex | | Descriptor: | MYOSIN A TAIL DOMAIN INTERACTING PROTEIN, MYOSIN-A | | Authors: | Salgado, P.S, Douse, C.H, Simpson, P.J, Thomas, J.C, Holder, A.A, Tate, E.W, Cota, E. | | Deposit date: | 2012-03-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Regulation of the Plasmodium Motor Complex: Phosphorylation of Myosin a Tail Interacting Protein (Mtip) Loosens its Grip on Myoa
J.Biol.Chem., 287, 2012
|
|
4AN0
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND INHIBITOR IC-3 | | Descriptor: | (2S)-1-[1-(4-phenylbutanoyl)-L-prolyl]pyrrolidine-2-carbonitrile, GLYCEROL, PROLYL ENDOPEPTIDASE | | Authors: | Kaszuba, K, Rog, T, Danne, R, Canning, P, Fulop, V, Juhasz, T, Szeltner, Z, St-Pierre, J.F, Garcia-Horsman, A, Mannisto, P.T, Karttunen, M, Hokkanen, J, Bunker, A. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular dynamics, crystallography and mutagenesis studies on the substrate gating mechanism of prolyl oligopeptidase.
Biochimie, 94, 2012
|
|
5I0U
 
 | |
2H8R
 
 | | Hepatocyte Nuclear Factor 1b bound to DNA: MODY5 Gene Product | | Descriptor: | 5'-D(*CP*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*TP*CP*AP*CP*CP*AP*G)-3', 5'-D(*GP*CP*TP*GP*GP*TP*GP*AP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3', Hepatocyte nuclear factor 1-beta | | Authors: | Lu, P, Rha, G.B, Chi, Y.I. | | Deposit date: | 2006-06-07 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of disease-causing mutations in hepatocyte nuclear factor 1beta.
Biochemistry, 46, 2007
|
|
2VH6
 
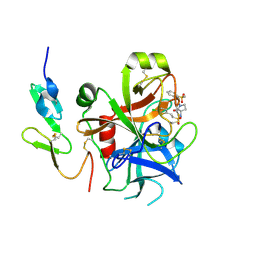 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with biaryl P4 motifs | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-{(3S)-1-[3-fluoro-2'-(methylsulfonyl)biphenyl-4-yl]-2-oxopyrrolidin-3-yl}ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN | | Authors: | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Chung, C.W, Convery, M.A, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A.N, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E. | | Deposit date: | 2007-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Biaryl P4 Motifs.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5LF9
 
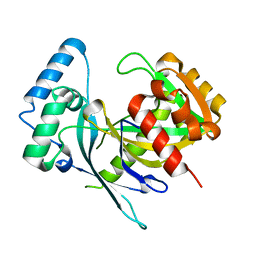 | | Crystal structure of human NUDT22 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 22 | | Authors: | Tallant, C, Siejka, P, Mathea, S, Shrestha, L, Krojer, T, Srikannathasan, V, Elkins, J.M, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human NUDT22
To Be Published
|
|
2H4I
 
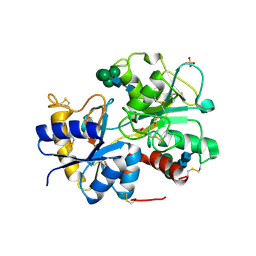 | | Crystal structure of the complex of proteolytically produced C-terminal half of bovine lactoferrin with lactose at 2.55 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem kumar, R, Sinha, M, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2006-05-24 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the complex of proteolytically produced C-terminal half of bovine lactoferrin with lactose at 2.55 A resolution
To be Published
|
|
4BQP
 
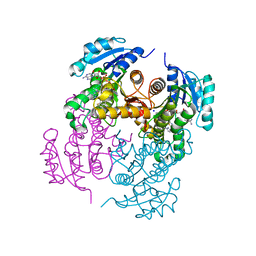 | | Mtb InhA complex with Methyl-thiazole compound 7 | | Descriptor: | (1S)-1-(5-{[1-(2,6-DIFLUOROBENZYL)-1H-PYRAZOL-3-YL]AMINO}-1,3,4-THIADIAZOL-2-YL)-1-(4-METHYL-1,3-THIAZOL-2-YL)ETHANOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Read, J.A, Gingell, H, Madhavapeddi, P, Shirude, P.S. | | Deposit date: | 2013-05-31 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Methyl-Thiazoles: A Novel Mode of Inhibition with the Potential to Develop Novel Inhibitors Targeting Inha in Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
7MVT
 
 | | Crystal structure of the Chaetomium thermophilum Nup192-Nic96 complex (Nup192 residues 185-1756; Nic96 residues 187-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP192 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
5KPO
 
 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[3-(4-ethyl-3-oxidanylidene-piperazin-1-yl)carbonyl-4-fluoranyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Yao, H.P, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
2HA2
 
 | | Crystal structure of mouse acetylcholinesterase complexed with succinylcholine | | Descriptor: | 2,2'-[(1,4-DIOXOBUTANE-1,4-DIYL)BIS(OXY)]BIS(N,N,N-TRIMETHYLETHANAMINIUM), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bourne, Y, Radic, Z, Sulzenbacher, G, Kim, E, Taylor, P, Marchot, P. | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate and product trafficking through the active center gorge of acetylcholinesterase analyzed by crystallography and equilibrium binding
J.Biol.Chem., 281, 2006
|
|
2V0T
 
 | | The A178L mutation in the C-terminal hinge of the flexible loop-6 of triosephosphate isomerase (TIM) induces a more closed conformation of this hinge region in dimeric and monomeric TIM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE GLYCOSOMAL | | Authors: | Alahuhta, M, Casteleijn, M.G, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-05-18 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies show that the A178L mutation in the C-terminal hinge of the catalytic loop-6 of triosephosphate isomerase (TIM) induces a closed-like conformation in dimeric and monomeric TIM.
Acta Crystallogr. D Biol. Crystallogr., 64, 2008
|
|
8PTL
 
 | | human PHOX2B C-terminal domain including the polyA fragment at 278K | | Descriptor: | Paired mesoderm homeobox protein 2B | | Authors: | Anton, R, Trevino, M.A, Pantoja-Uceda, D, Felix, S, Babu, M, Cabrita, E.J, Zweckstetter, M, Tinnefeld, P, Vera, A.M, Oroz, J. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions.
Nat Commun, 15, 2024
|
|
2HBN
 
 | |
5LK8
 
 | |
4WZY
 
 | | Structure of mycobacterial maltokinase, the missing link in the essential GlgE-pathway (ATP complex) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Maltokinase | | Authors: | Fraga, J, Empadinhas, N, Pereira, P.J.B, Macedo-Ribeiro, S. | | Deposit date: | 2014-11-20 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of mycobacterial maltokinase, the missing link in the essential GlgE-pathway.
Sci Rep, 5, 2015
|
|
3BBU
 
 | |
