4D0X
 
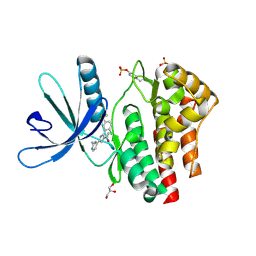 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 5-(2-aminopyrimidin-4-yl)-2-[2-chloro-5-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, GLYCEROL, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Canevari, G, Fasolini, M, Bertrand, J, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
2LIU
 
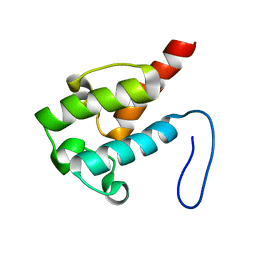 | | NMR structure of holo-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
4D1A
 
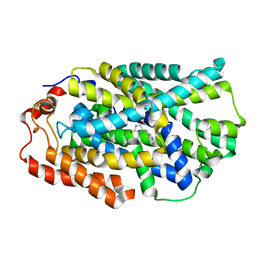 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | Descriptor: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D2O
 
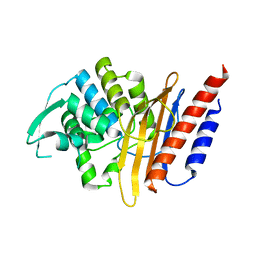 | | Crystal structure of the class A extended-spectrum beta-lactamase PER- 2 | | Descriptor: | PER-2 BETA-LACTAMASE | | Authors: | Power, P, Herman, R, Ruggiero, M, Kerff, F, Galleni, M, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2014-05-12 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Extended-Spectrum Beta-Lactamase Per- 2 and Insights Into the Role of Specific Residues in the Interaction with Beta-Lactams and Beta-Lactamase Inhibitors.
Antimicrob.Agents Chemother., 58, 2014
|
|
6MED
 
 | | Crystal structure of broadly neutralizing antibody HEPC3 | | Descriptor: | SULFATE ION, antibody HEPC3 Heavy Chain, antibody HEPC3 Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.
Cell Host Microbe, 24, 2018
|
|
4D82
 
 | | Metallosphera sedula Vps4 crystal structure | | Descriptor: | AAA ATPase, central domain protein, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Caillat, C, Macheboeuf, P, Wu, Y, McCarthy, A.A, Boeri-Erba, E, Effantin, G, Gottlinger, H.G, Weissenhorn, W, Renesto, P. | | Deposit date: | 2014-12-02 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Asymmetric Ring Structure of Vps4 Required for Escrt-III Disassembly.
Nat.Commun., 6, 2015
|
|
4D1B
 
 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | Descriptor: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
6MEM
 
 | |
1S0P
 
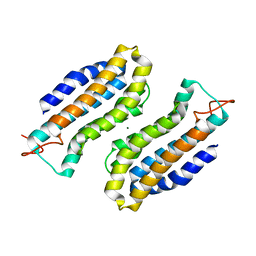 | | Structure of the N-Terminal Domain of the Adenylyl Cyclase-Associated Protein (CAP) from Dictyostelium discoideum. | | Descriptor: | Adenylyl cyclase-associated protein, MAGNESIUM ION | | Authors: | Ksiazek, D, Brandstetter, H, Israel, L, Bourenkov, G.P, Katchalova, G, Janssen, K.P, Bartunik, H.D, Noegel, A.A, Schleicher, M, Holak, T.A. | | Deposit date: | 2004-01-01 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | STRUCTURE OF THE N-TERMINAL DOMAIN OF THE ADENYLYL
CYCLASE-ASSOCIATED PROTEIN (CAP) FROM DICTYOSTELIUM DISCOIDEUM
Structure, 11, 2003
|
|
2LG2
 
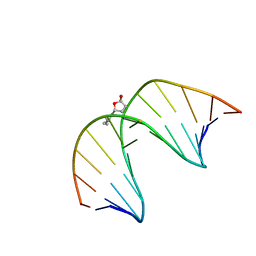 | | Structure of the duplex containing HNE derived (6S,8R,11S) N2-dG cyclic hemiacetal when placed opposite dT | | Descriptor: | (2R,5S)-5-pentyltetrahydrofuran-2-ol, DNA (5'-D(*GP*CP*TP*AP*GP*CP*GP*AP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*GP*CP*TP*AP*GP*C)-3') | | Authors: | Huang, H, Wang, H, Kozekova, A, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ring-chain tautomerization of trans-4-hydroxynonenal derived (6S,8R,11S) gamma-hydroxy-1,N2-propano-deoxyguanosine adduct when placed opposite 2'-deoxythymidine in duplex
To be Published
|
|
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1MJI
 
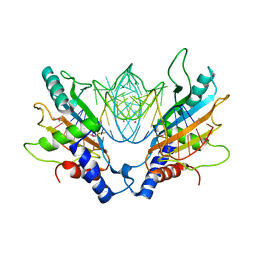 | | DETAILED ANALYSIS OF RNA-PROTEIN INTERACTIONS WITHIN THE BACTERIAL RIBOSOMAL PROTEIN L5/5S RRNA COMPLEX | | Descriptor: | 50S ribosomal protein L5, 5S rRNA fragment, MAGNESIUM ION, ... | | Authors: | Perederina, A, Nevskaya, N, Nikonov, O, Nikulin, A, Dumas, P, Yao, M, Tanaka, I, Garber, M, Gongadze, G, Nikonov, S. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detailed analysis of RNA-protein interactions within the bacterial ribosomal
protein L5/5S rRNA complex
RNA, 8, 2002
|
|
2QWU
 
 | |
2QXJ
 
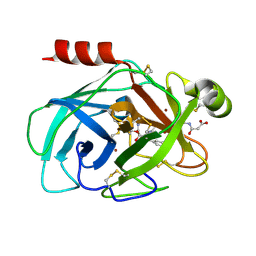 | | Crystal Structure of Human Kallikrein 7 in Complex with Suc-Ala-Ala-Pro-Phe-chloromethylketone and Copper | | Descriptor: | COPPER (II) ION, Kallikrein-7, N-(3-carboxypropanoyl)-L-alanyl-L-alanyl-N-[(2S,3S)-4-chloro-3-hydroxy-1-phenylbutan-2-yl]-L-prolinamide | | Authors: | Debela, M, Hess, P, Magdolen, V, Schechter, N.M, Bode, W, Goettig, P. | | Deposit date: | 2007-08-11 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chymotryptic specificity determinants in the 1.0 A structure of the zinc-inhibited human tissue kallikrein 7.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QY0
 
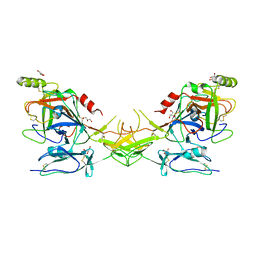 | | Active dimeric structure of the catalytic domain of C1r reveals enzyme-product like contacts | | Descriptor: | Complement C1r subcomponent, GLYCEROL | | Authors: | Kardos, J, Harmat, V, Pallo, A, Barabas, O, Szilagyi, K, Graf, L, Naray-Szabo, G, Goto, Y, Zavodszky, P, Gal, P. | | Deposit date: | 2007-08-13 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: Structure of the active catalytic region of C1r.
Mol.Immunol., 45, 2008
|
|
2R09
 
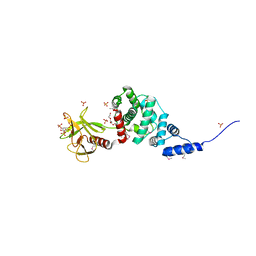 | | Crystal Structure of Autoinhibited Form of Grp1 Arf GTPase Exchange Factor | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Cytohesin-3, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ... | | Authors: | DiNitto, J.P, Delprato, A, Gabe Lee, M.T, Cronin, T.C, Huang, S, Guilherme, A, Czech, M.P, Lambright, D.G. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis and Mechanism of Autoregulation in 3-Phosphoinositide-Dependent Grp1 Family Arf GTPase Exchange Factors.
Mol.Cell, 28, 2007
|
|
4FCP
 
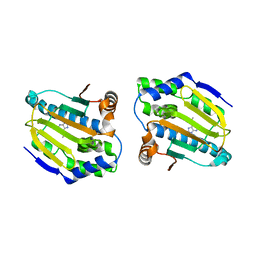 | | Targetting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo [2,3-d] pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | Descriptor: | Heat shock protein HSP 90-alpha, N,N-dimethyl-7H-purin-6-amine | | Authors: | Davies, N.G.M, Browne, H, Davies, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
3CY4
 
 | | Crystal Structure cation-dependent mannose 6-phosphate receptor at pH 7.4 | | Descriptor: | Cation-dependent mannose-6-phosphate receptor, GLYCEROL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Olson, L.J, Hindsgaul, O, Dahms, N.M, Kim, J.-J.P. | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-13 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Insights into the Mechanism of pH-dependent Ligand Binding and Release by the Cation-dependent Mannose 6-Phosphate Receptor.
J.Biol.Chem., 283, 2008
|
|
6LZQ
 
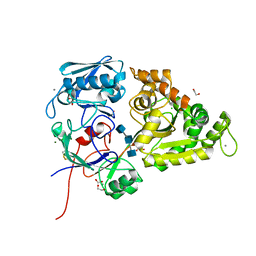 | | Chitin-specific solute binding protein from Vibrio harveyi in complex with chitotriose. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kitaoku, Y, Ubonbal, P, Tran, L.T, Robinson, R.C, Suginta, W. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural model for (GlcNAc) 2 translocation via a periplasmic chitooligosaccharide-binding protein from marine Vibrio bacteria.
J.Biol.Chem., 297, 2021
|
|
6M1X
 
 | |
1WTX
 
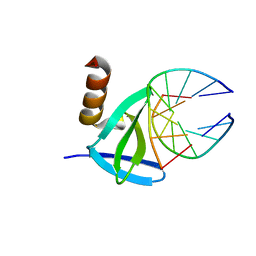 | | Hyperthermophile chromosomal protein SAC7D single mutant V26A in complex with DNA GTAATTAC | | Descriptor: | 5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Chen, C.-Y, Ko, T.-P, Lin, T.-W, Chou, C.-C, Chen, C.-J, Wang, A.H.-J. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d
NUCLEIC ACIDS RES., 33, 2005
|
|
4FFB
 
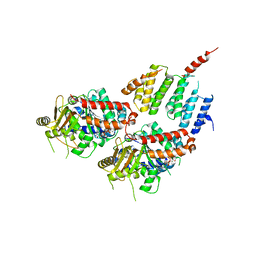 | | A TOG:alpha/beta-tubulin Complex Structure Reveals Conformation-Based Mechanisms For a Microtubule Polymerase | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | Authors: | Ayaz, P, Ye, X, Huddleston, P, Brautigam, C.A, Rice, L.M. | | Deposit date: | 2012-05-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | A TOG: alpha beta-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase.
Science, 337, 2012
|
|
2LT7
 
 | | Solution NMR structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2L04
 
 | | The Solution Structure of the C-terminal Ig-like Domain of the Bacteriophage Lambda Tail Tube Protein | | Descriptor: | Major tail protein V | | Authors: | Pell, L.G, Gasmi-Seabrook, G.M.C, Donaldson, L.W, Howell, P, Davidson, A.R, Maxwell, K.L. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of the C-Terminal Ig-like Domain of the Bacteriophage l Tail Tube Protein.
J.Mol.Biol., 403, 2010
|
|
2QG1
 
 | | Crystal structure of the 11th PDZ domain of MPDZ (MUPP1) | | Descriptor: | 1,2-ETHANEDIOL, Multiple PDZ domain protein | | Authors: | Papagrigoriou, E, Salah, E, Phillips, C, Savitsky, P, Boisguerin, P, Oschkinat, H, Gileadi, C, Yang, X, Elkins, J.M, Ugochukwu, E, Bunkoczi, G, Uppenberg, J, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the 11th PDZ domain of MPDZ (MUPP1).
To be Published
|
|
