6SYZ
 
 | | Crystal structure of YTHDC1 with fragment 1 (DHU_DC1_141) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methylthieno[3,2-d]pyrimidin-4-amine | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
9DN4
 
 | | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N | | Descriptor: | CHLORIDE ION, FAB BL3-6S97N HEAVY CHAIN, FAB BL3-6S97N LIGHT CHAIN, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Hegde, S, Wang, J. | | Deposit date: | 2024-09-16 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a SARS-CoV-2 20-mer RNA in complex with FAB BL3-6S97N
To be published
|
|
8CZX
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, TETRAETHYLENE GLYCOL, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, ... | | Authors: | Machen, A.J, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
1TK4
 
 | | Crystal structure of russells viper phospholipase A2 in complex with a specifically designed tetrapeptide Ala-Ile-Arg-Ser at 1.1 A resolution | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa, SULFATE ION, Tetrapeptide Ala-Ile-Arg-Ser | | Authors: | Singh, N, Bilgrami, S, Somvanshi, R.K, Sharma, S, Dey, S, Perbandt, M, Betzel, C, Kaur, P, Singh, T.P. | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of russells viper phospholipase A2 with a specifically designed tetrapeptide Ala-Ile-Arg-Ser at 1.1 A resolution
TO BE PUBLISHED
|
|
8CZW
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 15d | | Descriptor: | 3C-like proteinase, TETRAETHYLENE GLYCOL, [(1~{S},2~{R})-2-cyclohexylcyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, ... | | Authors: | Machen, A.J, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
2IVB
 
 | | SITE DIRECTED MUTAGENESIS OF KEY RESIDUES INVOLVED IN THE CATALYTIC MECHANISM OF CYANASE | | Descriptor: | AZIDE ION, CHLORIDE ION, CYANATE HYDRATASE, ... | | Authors: | Guilloton, M, Walsh, M.A, Joachimiak, A, Anderson, P.M. | | Deposit date: | 2006-06-09 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Twin Set of Low Pka Arginines Ensures the Concerted Acid Base Catalytic Mechanism of Cyanase
To be Published
|
|
7WOJ
 
 | | Crystal structure of HSA-Myr complex soaked with cisplatin for one week | | Descriptor: | Albumin, Cisplatin, MYRISTIC ACID | | Authors: | Chen, S.L, Yuan, C, Jiang, L.G, Luo, Z.P, Huang, M.D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystallographic analysis of interaction between cisplatin and human serum albumin: Effect of fatty acid.
Int.J.Biol.Macromol., 216, 2022
|
|
5AOH
 
 | | Crystal Structure of CarF | | Descriptor: | POTASSIUM ION, Spore coat protein CotH | | Authors: | Tichy, E.M, Hardwick, S.W, Luisi, B.F, C Salmond, G.P. | | Deposit date: | 2015-09-10 | | Release date: | 2017-01-25 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 angstrom resolution crystal structure of the carbapenem intrinsic resistance protein CarF.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2IVQ
 
 | |
5YXZ
 
 | | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI484 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-8-fluoranyl-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, ... | | Authors: | Swaminathan, S, Thakur, M.K, Kandan, S, Gautam, A, Kanavalli, M, Simhadri, P, Gosu, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI484
To Be Published
|
|
2Y75
 
 | | The Structure of CymR (YrzC) the Global Cysteine Regulator of B. subtilis | | Descriptor: | CHLORIDE ION, HTH-TYPE TRANSCRIPTIONAL REGULATOR CYMR, SULFATE ION | | Authors: | Shepard, W, Soutourina, O, Courtois, E, England, P, Haouz, A, Martin-Verstraete, I. | | Deposit date: | 2011-01-28 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights Into the Rrf2 Repressor Family - the Structure of Cymr, the Global Cysteine Regulator of Bacillus Subtilis.
FEBS J., 278, 2011
|
|
5YY1
 
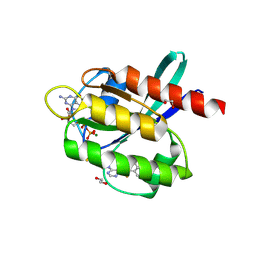 | | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI739 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[6-chloranyl-8-fluoranyl-7-[2-(trifluoromethyl)phenyl]quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, ... | | Authors: | Swaminathan, S, Thakur, M.K, Kandan, S, Gautam, A, Kanavalli, M, Simhadri, P, Gosu, R. | | Deposit date: | 2017-12-07 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Co-crystal Structure of KRAS (G12C) covalently bound with Quinazoline based inhibitor JBI739
To Be Published
|
|
6JD1
 
 | | Cryo-EM Structure of Sulfolobus solfataricus ketol-acid reductoisomerase (Sso-KARI) in complex with Mg2+, NADH, and CPD at pH7.5 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MAGNESIUM ION, Putative ketol-acid reductoisomerase 2, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, K.F, Huang, C.H, Lin, B.L, Ko, T.P, Hsieh, D.L, Tsai, M.D. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Use of Cryo-EM To Uncover Structural Bases of pH Effect and Cofactor Bispecificity of Ketol-Acid Reductoisomerase.
J. Am. Chem. Soc., 141, 2019
|
|
2Y7J
 
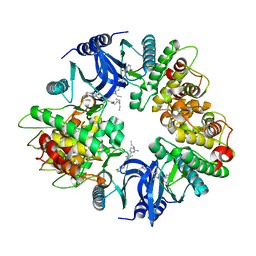 | | Structure of human phosphorylase kinase, gamma 2 | | Descriptor: | N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, PHOSPHORYLASE B KINASE GAMMA CATALYTIC CHAIN, TESTIS/LIVER ISOFORM | | Authors: | Muniz, J.R.C, Shrestha, A, Savitsky, P, Wang, J, Rellos, P, Fedorov, O, Burgess-Brown, N, Brenner, B, Berridge, G, Elkins, J.M, Krojer, T, Vollmar, M, Che, K.H, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Phosphorylase Kinase, Gamma 2
To be Published
|
|
1TJW
 
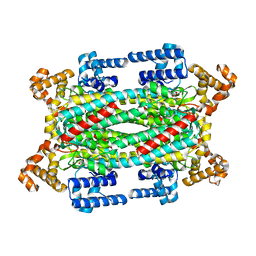 | | Crystal Structure of T161D Duck Delta 2 Crystallin Mutant with bound argininosuccinate | | Descriptor: | ARGININOSUCCINATE, Delta crystallin II | | Authors: | Sampaleanu, L.M, Codding, P.W, Lobsanov, Y.D, Tsai, M, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2004-06-07 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of duck delta2 crystallin mutants provide insight into the role of Thr161 and the 280s loop in catalysis
Biochem.J., 384, 2004
|
|
6T01
 
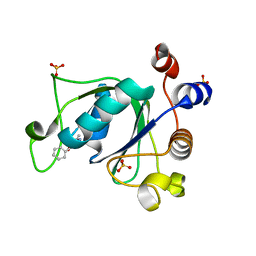 | | Crystal structure of YTHDC1 with fragment 13 (DHU_DC1_153) | | Descriptor: | 6-methyl-5-nitro-4-phenyl-1~{H}-pyrimidin-2-one, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
1TFV
 
 | | CRYSTAL STRUCTURE OF A BUFFALO SIGNALING GLYCOPROTEIN (SPB-40) SECRETED DURING INVOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, mammary gland protein 40 | | Authors: | Bilgrami, S, Saravanan, K, Yadav, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CRYSTAL STRUCTURE OF A BUFFALO SIGNALING GLYCOPROTEIN (SPB-40)
SECRETED DURING INVOLUTION
To be Published
|
|
6T0A
 
 | | Crystal structure of YTHDC1 with fragment 25 (PSI_DC1_005) | | Descriptor: | (~{R})-azanyl(pyridin-3-yl)methanethiol, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6SIB
 
 | |
2IW6
 
 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2-CYCLIN A COMPLEXED WITH A BISANILINOPYRIMIDINE INHIBITOR | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, MAGNESIUM ION, ... | | Authors: | Pratt, D.J, Bentley, J, Jewsbury, P, Boyle, F.T, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2006-06-26 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the Determinants of Cyclin-Dependent Kinase 2 and Cyclin-Dependent Kinase 4 Inhibitor Selectivity.
J.Med.Chem., 49, 2006
|
|
2LZV
 
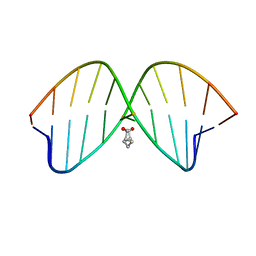 | | DNA duplex containing mispair-aligned O4U-heptylene-O4U interstrand cross-link | | Descriptor: | DNA (5'-D(*CP*GP*AP*AP*AP*UP*TP*TP*TP*CP*G)-3'), HEPTANE | | Authors: | Denisov, A.Y, McManus, F.P, Noronha, A.M, Wilds, C.J. | | Deposit date: | 2012-10-11 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of interstrand cross-link repair by O 6 -alkylguanine DNA alkyltransferase.
Org.Biomol.Chem., 15, 2017
|
|
7F6T
 
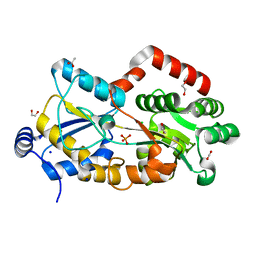 | |
1TBC
 
 | |
2Y7H
 
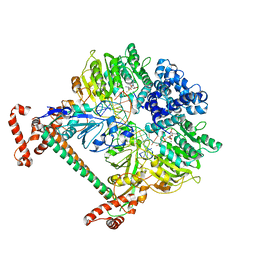 | | Atomic model of the DNA-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | 5'-D(*GP*TP*TP*CP*AP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*GP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*CP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*TP*GP*AP*AP*C)-3', S-ADENOSYLMETHIONINE, ... | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
7WBN
 
 | | PDB structure of RevCC | | Descriptor: | RevCC | | Authors: | Han, S, Kim, D, Kaur, M, Lim, Y.B, Barnwal, R.P. | | Deposit date: | 2021-12-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pseudo-Isolated alpha-Helix Platform for the Recognition of Deep and Narrow Targets.
J.Am.Chem.Soc., 144, 2022
|
|
