1KRC
 
 | |
8QPY
 
 | |
1YYM
 
 | | crystal structure of F23, a scorpion-toxin mimic of CD4, in complex with HIV-1 YU2 gp120 envelope glycoprotein and anti-HIV-1 antibody 17b | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Exterior membrane glycoprotein(GP120),Exterior membrane glycoprotein(GP120),Exterior membrane glycoprotein(GP120), ... | | Authors: | Huang, C.C, Stricher, F, Martin, L, Decker, J.M, Majeed, S, Barthe, P, Hendrickson, W.A, Robinson, J, Roumestand, C, Sodroski, J, Wyatt, R, Shaw, G.M, Vita, C, Kwong, P.D. | | Deposit date: | 2005-02-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth.
Structure, 13, 2005
|
|
5U3V
 
 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 6 | | Descriptor: | 6-[2-({cyclopentyl[4-(furan-3-yl)benzene-1-carbonyl]amino}methyl)phenoxy]hexanoic acid, DI(HYDROXYETHYL)ETHER, Peroxisome proliferator-activated receptor delta, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1ZNF
 
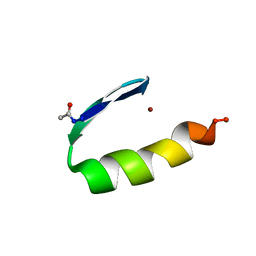 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SINGLE ZINC FINGER DNA-BINDING DOMAIN | | Descriptor: | 31ST ZINC FINGER FROM XFIN, ZINC ION | | Authors: | Lee, M.S, Gippert, G.P, Soman, K.V, Case, D.A, Wright, P.E. | | Deposit date: | 1989-09-25 | | Release date: | 1991-07-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of a single zinc finger DNA-binding domain.
Science, 245, 1989
|
|
4XUG
 
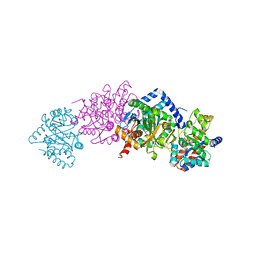 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with 2-({[4-(Trifluoromethoxy)Phenyl]Sulfonyl}Amino)Ethyl Dihydrogen Phosphate (F9F) inhibitor in the alpha site and ammonium ion in the metal coordination site. | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, AMMONIUM ION, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2015-01-25 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with 2-({[4-(Trifluoromethoxy)Phenyl]Sulfonyl}Amino)Ethyl Dihydrogen Phosphate (F9F) inhibitor in the alpha site and ammonium ion in the metal coordination site.
To Be Published
|
|
7KXS
 
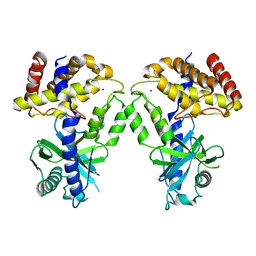 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2020-12-04 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2X7T
 
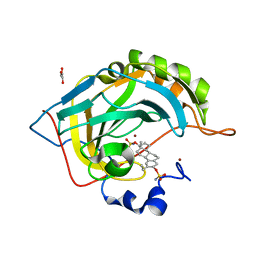 | | Structures of human carbonic anhydrase II inhibitor complexes reveal a second binding site for steroidal and non-steroidal inhibitors. | | Descriptor: | (9BETA,13ALPHA,14BETA,17ALPHA)-2-ETHYLESTRA-1(10),2,4-TRIENE-3,17-DIYL DISULFAMATE, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Cozier, G.E, Leese, M.P, Lloyd, M.D, Baker, M.D, Thiyagarajan, N, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures of Human Carbonic Anhydrase II/Inhibitor Complexes Reveal a Second Binding Site for Steroidal and Non-Steroidal Inhibitors.
Biochemistry, 49, 2010
|
|
5U4X
 
 | | Coactivator-associated arginine methyltransferase 1 with TP-064 | | Descriptor: | Histone-arginine methyltransferase CARM1, N-methyl-N-[(2-{1-[2-(methylamino)ethyl]piperidin-4-yl}pyridin-4-yl)methyl]-3-phenoxybenzamide, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | DONG, A, ZENG, H, Saikatendu, K.S, STONE, H, WALKER, J.R, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-06 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma.
Oncotarget, 9, 2018
|
|
5MO7
 
 | | Crystal Structure of CK2alpha with N-(3-(((2-chloro-[1,1'-biphenyl]-4-yl)methyl)amino)propyl)methanesulfonamide bound | | Descriptor: | 3-[(3-chloranyl-4-phenyl-phenyl)methylamino]propanamide, Casein kinase II subunit alpha, PHOSPHATE ION | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-13 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
5MOV
 
 | | Crystal structure of Ck2alpha with ZT0633 bound | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Casein kinase II subunit alpha | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-14 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
2X23
 
 | | crystal structure of M. tuberculosis InhA inhibited by PT70 | | Descriptor: | 5-HEXYL-2-(2-METHYLPHENOXY)PHENOL, DIMETHYL SULFOXIDE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], ... | | Authors: | Luckner, S.R, Liu, N, am Ende, C.W, Tonge, P.J, Kisker, C. | | Deposit date: | 2010-01-10 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | A Slow, Tight Binding Inhibitor of Inha, the Enoyl-Acyl Carrier Protein Reductase from Mycobacterium Tuberculosis.
J.Biol.Chem., 285, 2010
|
|
5TS5
 
 | |
5MPV
 
 | |
3WCG
 
 | | The complex structure of TcSQS with ligand, BPH1344 | | Descriptor: | Farnesyltransferase, putative, hydrogen [(1R)-2-(3-pentadecyl-1H-imidazol-3-ium-1-yl)-1-phosphonoethyl]phosphonate | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
6GK2
 
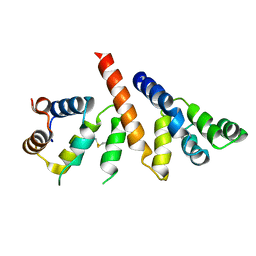 | | Helical reconstruction of BCL10 CARD and MALT1 DEATH DOMAIN complex | | Descriptor: | B-cell lymphoma/leukemia 10, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Schlauderer, F, Desfosses, A, Gutsche, I, Hopfner, K.P, Lammens, K. | | Deposit date: | 2018-05-18 | | Release date: | 2018-10-31 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture and regulation of BCL10-MALT1 filaments.
Nat Commun, 9, 2018
|
|
5JQH
 
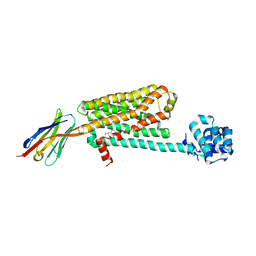 | | Structure of beta2 adrenoceptor bound to carazolol and inactive-state stabilizing nanobody, Nb60 | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, CHOLESTEROL, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Staus, D.P, Strachan, R.T, Manglik, A, Pani, B, Kahsai, A.W, Kim, T.H, Wingler, L.M, Ahn, S, Chatterjee, A, Masoudi, A, Kruse, A.C, Pardon, E, Steyaert, J, Weis, W.I, Prosser, R.S, Kobilka, B.K, Costa, T, Lefkowitz, R.J. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation.
Nature, 535, 2016
|
|
5M7Q
 
 | |
2WSY
 
 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
8R47
 
 | | AL amyloid fibril from the FOR010 light chain | | Descriptor: | lambda 3 immunoglobulin light chain fragment, residues 4-116 | | Authors: | Pfeiffer, P.B, Banerjee, S, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-11-13 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Light chain mutations contribute to defining the fibril morphology in systemic AL amyloidosis.
Nat Commun, 15, 2024
|
|
6GLJ
 
 | | Crystal structure of hMTH1 F27A in complex with TH scaffold 1 in the absence of acetate | | Descriptor: | 4-phenylpyrimidin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | hMTH1 F27A in complex with TH scaffold 1.
To Be Published
|
|
6GLO
 
 | |
6GMB
 
 | | Structure of human hydroxyacid oxidase 1 bound with FMN and glycolate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, GLYCOLIC ACID, ... | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Smee, C, Arrowsmith, C.H, Edwards, E, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-24 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with FMN and glycolate
To Be Published
|
|
5UGS
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT501 | | Descriptor: | 5-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]-2-(2-methylphenoxy)phenol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
1L7H
 
 | | Crystal structure of R292K mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
