1JR1
 
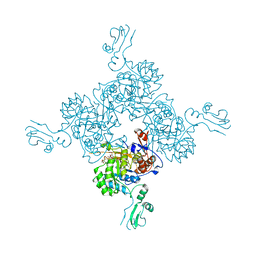 | | Crystal structure of Inosine Monophosphate Dehydrogenase in complex with Mycophenolic Acid | | Descriptor: | INOSINIC ACID, Inosine-5'-Monophosphate Dehydrogenase 2, MYCOPHENOLIC ACID, ... | | Authors: | Sintchak, M.D, Fleming, M.A, Futer, O, Raybuck, S.A, Chambers, S.P, Caron, P.R, Murcko, M.A, Wilson, K.P. | | Deposit date: | 2001-08-09 | | Release date: | 2001-09-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid.
Cell(Cambridge,Mass.), 85, 1996
|
|
4Z0Q
 
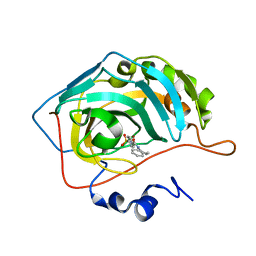 | | Carbonic anhydrase inhibitors: Design and synthesis of new heteroaryl-N-carbonylbenzenesulfonamides targeting druggable human carbonic anhydrase isoforms (hCA VII, hCA IX, and hCA XIV) | | Descriptor: | 4-(3,4-dihydroquinolin-1(2H)-ylcarbonyl)benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Pospisilova, K, Rezacova, P, Pachl, P. | | Deposit date: | 2015-03-26 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbonic anhydrase inhibitors: Design, synthesis and structural characterization of new heteroaryl-N-carbonylbenzenesulfonamides targeting druggable human carbonic anhydrase isoforms.
Eur.J.Med.Chem., 102, 2015
|
|
5AGU
 
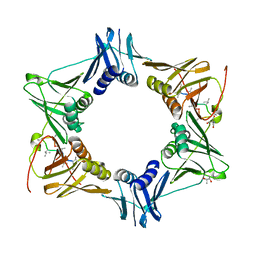 | | The sliding clamp of Mycobacterium tuberculosis in complex with a natural product. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA POLYMERASE III SUBUNIT BETA, ... | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
2XFD
 
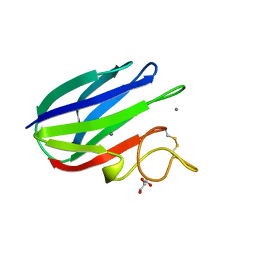 | | vCBM60 in complex with cellobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, GLYCEROL, ... | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
4Z1J
 
 | | Carbonic anhydrase inhibitors: Design and synthesis of new heteroaryl-N-carbonylbenzenesulfonamides targeting druggable human carbonic anhydrase isoforms (hCA VII, hCA IX, and hCA XIV) | | Descriptor: | 4-(3,4-dihydroisoquinolin-2(1H)-ylcarbonyl)benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Pospisilova, K, Rezacova, P, Pachl, P. | | Deposit date: | 2015-03-27 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Carbonic anhydrase inhibitors: Design, synthesis and structural characterization of new heteroaryl-N-carbonylbenzenesulfonamides targeting druggable human carbonic anhydrase isoforms.
Eur.J.Med.Chem., 102, 2015
|
|
7AAC
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with veliparib | | Descriptor: | (2R)-2-(7-carbamoyl-1H-benzimidazol-2-yl)-2-methylpyrrolidinium, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
6UTR
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with copper | | Descriptor: | ATP-dependent sacrificial sulfur transferase LarE, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Huizenga, K, Hausinger, R.P, Hu, J. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystallographic characterization of a tri-Asp metal-binding site at the three-fold symmetry axis of LarE.
Sci Rep, 10, 2020
|
|
5YTL
 
 | | Crystal structure of Geobacillus thermodenitrificans copper-containing nitrite reductase determined with an anaerobically manipulated crystal | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | Authors: | Fukuda, Y, Matsusaki, T, Tse, K.M, Mizohata, E, Murphy, M.E.P, Inoue, T. | | Deposit date: | 2017-11-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystallographic study of dioxygen chemistry in a copper-containing nitrite reductase from Geobacillus thermodenitrificans.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6S00
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-acetylneuraminic acid | | Descriptor: | GLYCEROL, N-acetyl-beta-neuraminic acid, TETRAETHYLENE GLYCOL, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
6S0E
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-Acetyl-2,3-dehydro-2-deoxyneuraminic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ACETATE ION, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-14 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
1T7J
 
 | | crystal structure of inhibitor amprenavir in complex with a multi-drug resistant variant of HIV-1 protease (L63P/V82T/I84V) | | Descriptor: | ACETATE ION, Pol Polyprotein, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | King, N.M, Prabu-Jeyabalan, M, Nalivaika, E.A, Wigerinck, P.B.T.P, De Bethune, M.-P, Schiffer, C.A. | | Deposit date: | 2004-05-10 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor
J.Med.Chem., 48, 2005
|
|
1HEJ
 
 | | C-terminal xylan binding domain from Cellulomonas fimi xylanase 11A | | Descriptor: | ENDO-1,4-BETA-XYLANASE D | | Authors: | Simpson, P.J, Hefang, X, Bolam, D.N, White, P, Hancock, S.M, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 2000-11-22 | | Release date: | 2001-05-10 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Evidence for Synergy between Family 2B Carbohydrate Binding Modules in Cellulomonas Fimi Xylanase 11A
Biochemistry, 40, 2001
|
|
1JBQ
 
 | | STRUCTURE OF HUMAN CYSTATHIONINE BETA-SYNTHASE: A UNIQUE PYRIDOXAL 5'-PHOSPHATE DEPENDENT HEMEPROTEIN | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meier, M, Janosik, M, Kery, V, Kraus, J.P, Burkhard, P. | | Deposit date: | 2001-06-06 | | Release date: | 2001-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein.
EMBO J., 20, 2001
|
|
5AE8
 
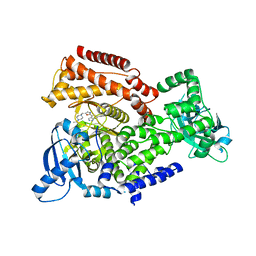 | | Crystal structure of mouse PI3 kinase delta in complex with GSK2269557 | | Descriptor: | 6-(1H-Indol-4-yl)-4-(5-{[4-(1-methylethyl)-1-piperazinyl]methyl}-1,3-oxazol-2-yl)-1H-indazole, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Down, K.D, Amour, A, Baldwin, I.R, Cooper, A.W.J, Deakin, A.M, Felton, L.M, Guntrip, S.B, Hardy, C, Harrison, Z.A, Jones, K.L, Jones, P, Keeling, S.E, Le, J, Livia, S, Lucas, F, Lunniss, C.J, Parr, N.J, Robinson, E, Rowland, P, Smith, S, Thomas, D.A, Vitulli, G, Washio, Y, Hamblin, N. | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Optimization of Novel Indazoles as Highly Potent and Selective Inhibitors of Phosphoinositide 3-Kinase Gamma for the Treatment of Respiratory Disease.
J.Med.Chem., 58, 2015
|
|
4L8T
 
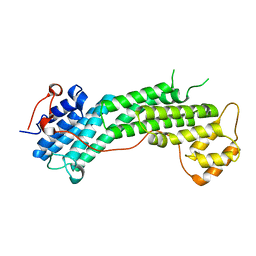 | | Structure of the Cargo Binding Domain from Human Myosin Vc | | Descriptor: | Unconventional myosin-Vc | | Authors: | Nascimento, A.F.Z, Tonoli, C.C.C, Trindade, D.M, Assis, L.H.P, Mahajan, P, Berridge, G, Krojer, T, Burgess-Brown, N, von Delft, F, Murakami, M.T. | | Deposit date: | 2013-06-17 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Insights into Functional Overlapping and Differentiation among Myosin V Motors.
J.Biol.Chem., 288, 2013
|
|
5AE9
 
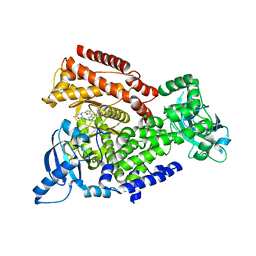 | | Crystal structure of mouse PI3 kinase delta in complex with GSK2292767 | | Descriptor: | N-[5-[4-(5-{[(2R,6S)-2,6-DIMETHYL-4-MORPHOLINYL]METHYL}-1,3-OXAZOL-2-YL)-1H-INDAZOL-6-YL]-2-(METHYLOXY)-3-PYRIDINYL]METHANESULFONAMIDE, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Down, K.D, Amour, A, Baldwin, I.R, Cooper, A.W.J, Deakin, A.M, Felton, L.M, Guntrip, S.B, Hardy, C, Harrison, Z.A, Jones, K.L, Jones, P, Keeling, S.E, Le, J, Livia, S, Lucas, F, Lunniss, C.J, Parr, N.J, Robinson, E, Rowland, P, Smith, S, Thomas, D.A, Vitulli, G, Washio, Y, Hamblin, N. | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Optimization of Novel Indazoles as Highly Potent and Selective Inhibitors of Phosphoinositide 3-Kinase Gamma for the Treatment of Respiratory Disease.
J.Med.Chem., 58, 2015
|
|
7AAD
 
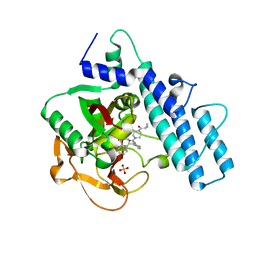 | | Crystal structure of the catalytic domain of human PARP1 in complex with olaparib | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
6HXA
 
 | | AntI from P. luminescens catalyses terminal polyketide shortening in the biosynthesis of anthraquinones | | Descriptor: | AntI, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Zhou, Q, Braeuer, A, Grammbitter, G, Schmalhofer, M, Saura, P, Adihou, H, Kaila, V.R.I, Groll, M, Bode, H. | | Deposit date: | 2018-10-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of polyketide shortening in anthraquinone biosynthesis ofPhotorhabdus luminescens.
Chem Sci, 10, 2019
|
|
2XWT
 
 | | CRYSTAL STRUCTURE OF THE TSH RECEPTOR IN COMPLEX WITH A BLOCKING TYPE TSHR AUTOANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, THYROID BLOCKING HUMAN AUTOANTIBODY K1-70 HEAVY CHAIN, ... | | Authors: | Sanders, J, Sanders, P, Young, S, Kabelis, K, Baker, S, Sullivan, A, Evans, M, Clark, J, Wilmot, J, Hu, X, Roberts, E, Powell, M, Nunez Miguel, R, Furmaniak, J, Rees Smith, B. | | Deposit date: | 2010-11-05 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Tsh Receptor (Tshr) Bound to a Blocking-Type Tshr Autoantibody.
J.Mol.Endocrinol., 46, 2011
|
|
4Z51
 
 | | High Resolution Human Septin 3 GTPase domain | | Descriptor: | MAGNESIUM ION, Neuronal-specific septin-3, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Valadares, N.F, Macedo, J.N, Leonardo, D.A, Brandao-Neto, J, Pereira, H.M, Matos, S.O, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2015-04-02 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A complete compendium of crystal structures for the human SEPT3 subgroup reveals functional plasticity at a specific septin interface
Iucrj, 2020
|
|
5D6T
 
 | | Crystal Structure of Aspergillus clavatus Sph3 in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, SPHERULIN-4, ... | | Authors: | Bamford, N.C, Little, D.J, Howell, P.L. | | Deposit date: | 2015-08-12 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Sph3 Is a Glycoside Hydrolase Required for the Biosynthesis of Galactosaminogalactan in Aspergillus fumigatus.
J.Biol.Chem., 290, 2015
|
|
6UW4
 
 | | Cryo-EM structure of human TRPV3 determined in lipid nanodisc | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7A7N
 
 | |
7A7L
 
 | | rsEGFP in the green-off state | | Descriptor: | Green fluorescent protein, TETRAETHYLENE GLYCOL | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7A7K
 
 | | rsEGFP in the green-on state | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
